过氧化氢诱导肺炎链球菌D39蛋白表达的比较蛋白质组学分析
IF 0.5
4区 生物学
Q4 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
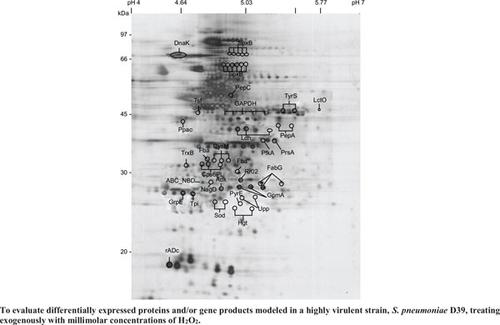
肺炎链球菌是人类呼吸道感染的主要原因。尽管缺乏抗氧化酶的活性,包括细胞色素、血红蛋白和过氧化物酶/过氧化氢酶,但在富氧环境中,这种细菌具有耐氧-厌氧生长的特性是保守的,具有很高的抗氧化作用。通过蛋白质组学分析,本研究的目的是评估用毫摩尔浓度h2o2外源处理的高毒力菌株肺炎链球菌D39模型中差异表达的蛋白质和/或基因产物。对于二维凝胶电泳(2-DE)分析,在固定化pH梯度为pH 4-7的一维等电聚焦下,通过SDS-PAGE在二维上分离表达最显著的蛋白。共检测到431个蛋白点,切除部分蛋白,酶切凝胶内胰蛋白酶,结合MALDI-TOF和LC-ESI-MS/MS进行质谱多肽定位和蛋白鉴定。利用质谱分析从2-DE中切除的斑点,选择的蛋白质斑点与各种数据库和MASCOT进行鉴定。通过与蛋白质组参考图的比较,外源2mm H2O2处理诱导了38个差异表达最大的蛋白,这些蛋白的增加和/或减少约1.4倍或更多,或具有多个表现可变pI值的异构体。鉴定的蛋白质被认为与肺炎球菌的发病机制和初级代谢等有关。这是第一个令人信服地记录肺炎链球菌在给定氧化条件下与病理生理适应相关的蛋白质组学信息,以及相应的潜在抗氧化机制的研究。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Comparative Proteomic Analysis of Hydrogen Peroxide-induced Protein Expression in Streptococcus pneumoniae D39
Streptococcus pneumoniae is a leading cause of human respiratory tract infection. Despite the lack
of activities of antioxidative enzymes, including cytochromes, hemoproteins, and peroxidases/catalases, traits conferring the
aerotolerant-anaerobic growth of this bacterium are conserved, with high efficacy of antioxidative actions, in an oxygen-rich
environment.
Through proteome analysis, this study’s intention was to evaluate differentially expressed proteins and/or gene
products modeled in a highly virulent strain, S. pneumoniae D39, exogenously-treated with millimolar concentrations of
H2O2.
For two-dimensional gel electrophoresis (2-DE) analysis, following one dimensional isoelectric focusing with an
immobilized pH gradient of pH 4-7, the most significantly mobilized proteins expressed were separated by SDS-PAGE in
the second dimension. With a total of 431 protein spots detected, certain proteins were excised, in-gel trypsin digested, and
analyzed by combination with MALDI-TOF and LC-ESI-MS/MS for mass spectrometric peptide mapping and protein
identification. Utilizing mass spectrometry analysis of spots excised from 2-DE, the selected protein spots were identified
with a variety of databases and MASCOT.
With the aid of comparisons to proteome reference maps, the most differentially expressed 38 proteins, those with
approximately 1.4-fold or more increase and/or decrease or with multiple isoforms exhibiting variable pI values, were
induced by treatment of exogenous 2 mM H2O2. The identified proteins were seen to be involved in pneumococcal
pathogenesis and primary metabolism, amongst others.
This is the first study to convincingly document proteomic information associated with pathophysiological
adaptation under the given oxidative conditions, and corresponding potential antioxidative mechanisms, in S. pneumoniae.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Current Proteomics
BIOCHEMICAL RESEARCH METHODS-BIOCHEMISTRY & MOLECULAR BIOLOGY
CiteScore
1.60
自引率
0.00%
发文量
25
审稿时长
>0 weeks
期刊介绍:
Research in the emerging field of proteomics is growing at an extremely rapid rate. The principal aim of Current Proteomics is to publish well-timed in-depth/mini review articles in this fast-expanding area on topics relevant and significant to the development of proteomics. Current Proteomics is an essential journal for everyone involved in proteomics and related fields in both academia and industry.
Current Proteomics publishes in-depth/mini review articles in all aspects of the fast-expanding field of proteomics. All areas of proteomics are covered together with the methodology, software, databases, technological advances and applications of proteomics, including functional proteomics. Diverse technologies covered include but are not limited to:
Protein separation and characterization techniques
2-D gel electrophoresis and image analysis
Techniques for protein expression profiling including mass spectrometry-based methods and algorithms for correlative database searching
Determination of co-translational and post- translational modification of proteins
Protein/peptide microarrays
Biomolecular interaction analysis
Analysis of protein complexes
Yeast two-hybrid projects
Protein-protein interaction (protein interactome) pathways and cell signaling networks
Systems biology
Proteome informatics (bioinformatics)
Knowledge integration and management tools
High-throughput protein structural studies (using mass spectrometry, nuclear magnetic resonance and X-ray crystallography)
High-throughput computational methods for protein 3-D structure as well as function determination
Robotics, nanotechnology, and microfluidics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


