好氧颗粒污泥中嗜麦芽窄食单胞菌AGS-1基因敲除CRISPR干扰系统的构建。
IF 3.9
2区 生物学
Q1 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
为了鉴定从好氧颗粒污泥中分离的嗜麦芽窄食单胞菌AGS-1中参与生物膜形成的附着基因的功能,需要一种有效的基因分子工具。我们在嗜麦芽窄食单胞菌AGS-1中开发了一个双质粒CRISPRi系统。一个质粒用l-阿拉伯糖诱导型启动子表达dCas9蛋白,另一个质粒含有与靶基因互补的sgRNA盒。在araC诱导型启动子的控制下,该系统表现出很少的基础表达泄漏和高度诱导表达,通过可逆敲低沉默内源性和外源性基因。与模板链(91倍)相比,该系统对非模板链上mCherry的表达实现了高达211倍的抑制。所开发的CRISPRi平台的实用性还通过抑制xanA和rpfF基因来表征。这两个基因的表达迅速减少,粘附能力下降,这表明任何一个基因的调节都是AGS-1菌株形成生物膜的重要因素。该系统还测试了同时沉默多个靶基因、整个操纵子或其部分转录抑制的能力。最后,CRISPRi的使用使我们能够剖析鞭毛生物合成中涉及的基因复杂性。总之,这些结果表明CRISPRi系统是一个简单、可行和可控的AGS-1菌株基因表达操作系统。本文章由计算机程序翻译,如有差异,请以英文原文为准。
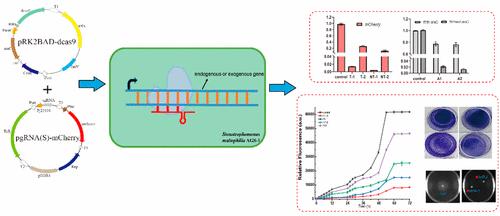
Construction of a CRISPR Interference System for Gene Knockdown in Stenotrophomonas maltophilia AGS-1 from Aerobic Granular Sludge
To identify the function of attachment genes involved in biofilm formation in Stenotrophomonas maltophilia AGS-1 isolated from aerobic granular sludge, an effective gene molecular tool is needed. We developed a two-plasmid CRISPRi system in Stenotrophomonas maltophilia AGS-1. One plasmid expressed dCas9 protein with the l-arabinose inducible promoter, and the other plasmid contained the sgRNA cassette complementary to the target gene. Under control of the araC-inducible promoter, this system exhibited little leaky basal expression and highly induced expression that silenced endogenous and exogenous genes with reversible knockdown. This system achieved up to 211-fold suppression for mCherry expression on the nontemplate strand compared to the template strand (91-fold). The utility of the developed CRISPRi platform was also characterized by suppressing the xanA and rpfF genes. The expression of these two genes was rapidly depleted and the adhesion ability decreased, which demonstrated that the modulation of either gene was an important factor for biofilm formation of the AGS-1 strain. The system also tested the ability to simultaneously silence transcriptional suppression of multiple targeted genes, an entire operon, or part of it. Lastly, the use of CRISPRi allowed us to dissect the gene intricacies involved in flagellar biosynthesis. Collectively, these results demonstrated that the CRISPRi system was a simple, feasible, and controllable manipulation system of gene expression in the AGS-1 strain.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
8.00
自引率
10.60%
发文量
380
审稿时长
6-12 weeks
期刊介绍:
The journal is particularly interested in studies on the design and synthesis of new genetic circuits and gene products; computational methods in the design of systems; and integrative applied approaches to understanding disease and metabolism.
Topics may include, but are not limited to:
Design and optimization of genetic systems
Genetic circuit design and their principles for their organization into programs
Computational methods to aid the design of genetic systems
Experimental methods to quantify genetic parts, circuits, and metabolic fluxes
Genetic parts libraries: their creation, analysis, and ontological representation
Protein engineering including computational design
Metabolic engineering and cellular manufacturing, including biomass conversion
Natural product access, engineering, and production
Creative and innovative applications of cellular programming
Medical applications, tissue engineering, and the programming of therapeutic cells
Minimal cell design and construction
Genomics and genome replacement strategies
Viral engineering
Automated and robotic assembly platforms for synthetic biology
DNA synthesis methodologies
Metagenomics and synthetic metagenomic analysis
Bioinformatics applied to gene discovery, chemoinformatics, and pathway construction
Gene optimization
Methods for genome-scale measurements of transcription and metabolomics
Systems biology and methods to integrate multiple data sources
in vitro and cell-free synthetic biology and molecular programming
Nucleic acid engineering.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


