Youjun Zhang, Roberto Natale, Adilson Pereira Domingues Júnior, Mitchell Rey Toleco, Beata Siemiatkowska, Norma Fàbregas, Alisdair R. Fernie
下载PDF
{"title":"植物中蛋白质-蛋白质相互作用的快速鉴定","authors":"Youjun Zhang, Roberto Natale, Adilson Pereira Domingues Júnior, Mitchell Rey Toleco, Beata Siemiatkowska, Norma Fàbregas, Alisdair R. Fernie","doi":"10.1002/cppb.20099","DOIUrl":null,"url":null,"abstract":"<p>Enzyme-enzyme interactions can be discovered by affinity purification mass spectrometry (AP-MS) under in vivo conditions. Tagged enzymes can either be transiently transformed into plant leaves or stably transformed into plant cells prior to AP-MS. The success of AP-MS depends on the levels and stability of the bait protein, the stability of the protein-protein interactions, and the efficiency of trypsin digestion and recovery of tryptic peptides for MS analysis. Unlike in-gel-digestion AP-MS, in which the gel is cut into pieces for several independent trypsin digestions, we uses a proteomics-based in-solution digestion method to directly digest the proteins on the beads following affinity purification. Thus, a single replicate within an AP-MS experiment constitutes a single sample for LC-MS measurement. In subsequent data analysis, normalized signal intensities can be processed to determine fold-change abundance (FC-A) scores by use of the SAINT algorithm embedded within the CRAPome software. Following analysis of co-sublocalization of “bait” and “prey,” we suggest considering only the protein pairs for which the intensities were more than 2% compared with the bait, corresponding to FC-A values of at least four within-biological replicates, which we recommend as minimum. If the procedure is faithfully followed, experimental assessment of enzyme-enzyme interactions can be carried out in Arabidopsis within 3 weeks (transient expression) or 5 weeks (stable expression). © 2019 The Authors.</p><p><b>Basic Protocol 1</b>: Gene cloning to the destination vectors</p><p><b>Alternate Protocol</b>: In-Fusion or Gibson gene cloning protocol</p><p><b>Basic Protocol 2</b>: Transformation of baits into the plant cell culture or plant leaf</p><p><b>Basic Protocol 3</b>: Affinity purification of protein complexes</p><p><b>Basic Protocol 4</b>: On-bead trypsin/LysC digestion and C18 column peptide desalting and concentration</p><p><b>Basic Protocol 5</b>: Data analysis and quality control</p>","PeriodicalId":10932,"journal":{"name":"Current protocols in plant biology","volume":"4 4","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2019-11-12","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://sci-hub-pdf.com/10.1002/cppb.20099","citationCount":"19","resultStr":"{\"title\":\"Rapid Identification of Protein-Protein Interactions in Plants\",\"authors\":\"Youjun Zhang, Roberto Natale, Adilson Pereira Domingues Júnior, Mitchell Rey Toleco, Beata Siemiatkowska, Norma Fàbregas, Alisdair R. Fernie\",\"doi\":\"10.1002/cppb.20099\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p>Enzyme-enzyme interactions can be discovered by affinity purification mass spectrometry (AP-MS) under in vivo conditions. Tagged enzymes can either be transiently transformed into plant leaves or stably transformed into plant cells prior to AP-MS. The success of AP-MS depends on the levels and stability of the bait protein, the stability of the protein-protein interactions, and the efficiency of trypsin digestion and recovery of tryptic peptides for MS analysis. Unlike in-gel-digestion AP-MS, in which the gel is cut into pieces for several independent trypsin digestions, we uses a proteomics-based in-solution digestion method to directly digest the proteins on the beads following affinity purification. Thus, a single replicate within an AP-MS experiment constitutes a single sample for LC-MS measurement. In subsequent data analysis, normalized signal intensities can be processed to determine fold-change abundance (FC-A) scores by use of the SAINT algorithm embedded within the CRAPome software. Following analysis of co-sublocalization of “bait” and “prey,” we suggest considering only the protein pairs for which the intensities were more than 2% compared with the bait, corresponding to FC-A values of at least four within-biological replicates, which we recommend as minimum. If the procedure is faithfully followed, experimental assessment of enzyme-enzyme interactions can be carried out in Arabidopsis within 3 weeks (transient expression) or 5 weeks (stable expression). © 2019 The Authors.</p><p><b>Basic Protocol 1</b>: Gene cloning to the destination vectors</p><p><b>Alternate Protocol</b>: In-Fusion or Gibson gene cloning protocol</p><p><b>Basic Protocol 2</b>: Transformation of baits into the plant cell culture or plant leaf</p><p><b>Basic Protocol 3</b>: Affinity purification of protein complexes</p><p><b>Basic Protocol 4</b>: On-bead trypsin/LysC digestion and C18 column peptide desalting and concentration</p><p><b>Basic Protocol 5</b>: Data analysis and quality control</p>\",\"PeriodicalId\":10932,\"journal\":{\"name\":\"Current protocols in plant biology\",\"volume\":\"4 4\",\"pages\":\"\"},\"PeriodicalIF\":0.0000,\"publicationDate\":\"2019-11-12\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://sci-hub-pdf.com/10.1002/cppb.20099\",\"citationCount\":\"19\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Current protocols in plant biology\",\"FirstCategoryId\":\"1085\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/cppb.20099\",\"RegionNum\":0,\"RegionCategory\":null,\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"Q1\",\"JCRName\":\"Agricultural and Biological Sciences\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols in plant biology","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cppb.20099","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q1","JCRName":"Agricultural and Biological Sciences","Score":null,"Total":0}
引用次数: 19
引用
批量引用
Rapid Identification of Protein-Protein Interactions in Plants
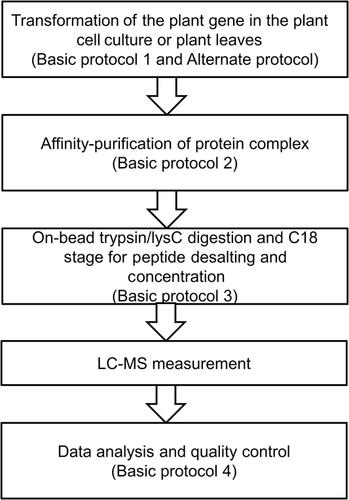
Enzyme-enzyme interactions can be discovered by affinity purification mass spectrometry (AP-MS) under in vivo conditions. Tagged enzymes can either be transiently transformed into plant leaves or stably transformed into plant cells prior to AP-MS. The success of AP-MS depends on the levels and stability of the bait protein, the stability of the protein-protein interactions, and the efficiency of trypsin digestion and recovery of tryptic peptides for MS analysis. Unlike in-gel-digestion AP-MS, in which the gel is cut into pieces for several independent trypsin digestions, we uses a proteomics-based in-solution digestion method to directly digest the proteins on the beads following affinity purification. Thus, a single replicate within an AP-MS experiment constitutes a single sample for LC-MS measurement. In subsequent data analysis, normalized signal intensities can be processed to determine fold-change abundance (FC-A) scores by use of the SAINT algorithm embedded within the CRAPome software. Following analysis of co-sublocalization of “bait” and “prey,” we suggest considering only the protein pairs for which the intensities were more than 2% compared with the bait, corresponding to FC-A values of at least four within-biological replicates, which we recommend as minimum. If the procedure is faithfully followed, experimental assessment of enzyme-enzyme interactions can be carried out in Arabidopsis within 3 weeks (transient expression) or 5 weeks (stable expression). © 2019 The Authors.
Basic Protocol 1 : Gene cloning to the destination vectors
Alternate Protocol : In-Fusion or Gibson gene cloning protocol
Basic Protocol 2 : Transformation of baits into the plant cell culture or plant leaf
Basic Protocol 3 : Affinity purification of protein complexes
Basic Protocol 4 : On-bead trypsin/LysC digestion and C18 column peptide desalting and concentration
Basic Protocol 5 : Data analysis and quality control

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


