基于自然语言处理(NLP)的特征提取技术在深度学习模型中预测分子的血脑屏障通透性
IF 2.8
4区 医学
Q3 CHEMISTRY, MEDICINAL
引用次数: 0
摘要
本文章由计算机程序翻译,如有差异,请以英文原文为准。
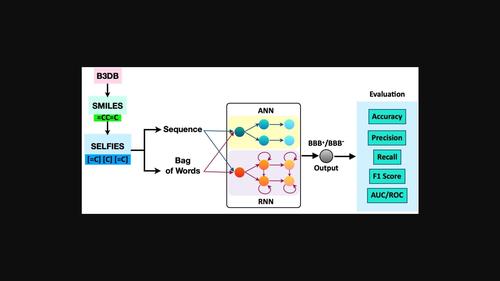
Natural‐Language Processing (NLP) based feature extraction technique in Deep‐Learning model to predict the Blood‐Brain‐Barrier permeability of molecules
Blood‐Brain‐Barrier (BBB) permeability is one of the critical factors in the success and failure of CNS drug development. The most accurate method of measuring BBB permeability involves clinical experiments, which are labour‐intensive and time‐consuming. Thus, numerous efforts were made to use artificial intelligence (AI) to predict molecules′ BBB permeability. Most of the previous models are based on calculated descriptors and molecular fingerprints. In the present work, we have developed an NLP‐based feature extraction technique in Deep‐Learning models to predict BBB permeability. We have used the B3DB database and generated SELFIES to extract features from the molecules. We have employed word level and N‐gram tokenization to represent words into numeric vectors. The extracted features were fed into several Artificial Neural Network (ANN) and Bi‐directional Long Short‐Term Memory (LSTM) models. The model, ANN‐10 built using ANN and 6‐gram tokenization, performed best on the independent test set. The accuracy, precision, recall, F1, specificity and AUC of ROC scores were found to be 0.89, 0.91, 0.91, 0.91, 0.85 and 0.90. Thus, the developed model can be used for the early screening of CNS drugs.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Molecular Informatics
CHEMISTRY, MEDICINAL-MATHEMATICAL & COMPUTATIONAL BIOLOGY
CiteScore
7.30
自引率
2.80%
发文量
70
审稿时长
3 months
期刊介绍:
Molecular Informatics is a peer-reviewed, international forum for publication of high-quality, interdisciplinary research on all molecular aspects of bio/cheminformatics and computer-assisted molecular design. Molecular Informatics succeeded QSAR & Combinatorial Science in 2010.
Molecular Informatics presents methodological innovations that will lead to a deeper understanding of ligand-receptor interactions, macromolecular complexes, molecular networks, design concepts and processes that demonstrate how ideas and design concepts lead to molecules with a desired structure or function, preferably including experimental validation.
The journal''s scope includes but is not limited to the fields of drug discovery and chemical biology, protein and nucleic acid engineering and design, the design of nanomolecular structures, strategies for modeling of macromolecular assemblies, molecular networks and systems, pharmaco- and chemogenomics, computer-assisted screening strategies, as well as novel technologies for the de novo design of biologically active molecules. As a unique feature Molecular Informatics publishes so-called "Methods Corner" review-type articles which feature important technological concepts and advances within the scope of the journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


