利用从全基因组关联研究中获得的 SNPs 的机器学习模型预测长春新碱诱发的周围神经病变的发病情况
IF 2.9
3区 医学
Q2 GENETICS & HEREDITY
引用次数: 1
摘要
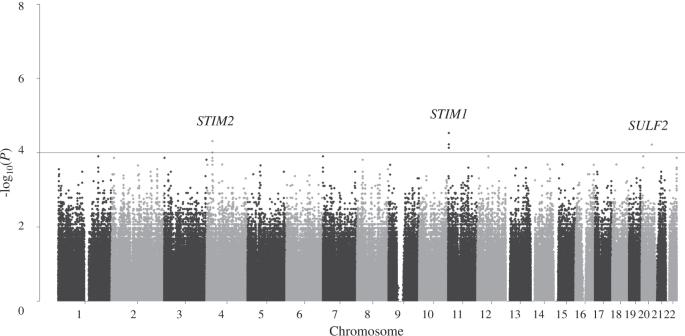
长春新碱治疗可能会导致周围神经病变。在这项研究中,我们利用全基因组关联研究(GWAS)确定了与长春新碱治疗导致的周围神经病变相关的基因,并利用基于遗传信息的机器学习构建了一个周围神经病变发生的预测模型。研究对象包括德岛大学医院血液科收治的 72 名接受长春新碱治疗的患者。使用 Illumina Asian Screening Array-24 套件对其中 56 例进行了基因分型,并对长春新碱导致的周围神经病变的发病进行了 GWAS 分析。通过对 16 个验证样本进行 Sanger 测序,确定了与外周神经病发病相关的前三个单核苷酸多态性 (SNP)。使用 R 软件包 "caret "进行了机器学习。56 个 GWAS 样本和 16 个验证样本分别作为训练集和测试集。使用随机森林、支持向量机、天真贝叶斯和神经网络算法构建了预测模型。根据 GWAS 的结果,rs2110179、rs7126100 和 rs2076549 与长春新碱用药导致的周围神经病变有关。利用这三个 SNPs 进行机器学习,构建了一个预测模型。使用支持向量机和神经网络对 rs2110179 和 rs2076549 的预测准确率高达 93.8%。因此,利用与长春新碱治疗相关的 SNPs 的机器学习预测模型可以有效地预测长春新碱治疗引起的周围神经病变。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A machine learning model using SNPs obtained from a genome-wide association study predicts the onset of vincristine-induced peripheral neuropathy
Vincristine treatment may cause peripheral neuropathy. In this study, we identified the genes associated with the development of peripheral neuropathy due to vincristine therapy using a genome-wide association study (GWAS) and constructed a predictive model for the development of peripheral neuropathy using genetic information-based machine learning. The study included 72 patients admitted to the Department of Hematology, Tokushima University Hospital, who received vincristine. Of these, 56 were genotyped using the Illumina Asian Screening Array-24 Kit, and a GWAS for the onset of peripheral neuropathy caused by vincristine was conducted. Using Sanger sequencing for 16 validation samples, the top three single nucleotide polymorphisms (SNPs) associated with the onset of peripheral neuropathy were determined. Machine learning was performed using the statistical software R package “caret”. The 56 GWAS and 16 validation samples were used as the training and test sets, respectively. Predictive models were constructed using random forest, support vector machine, naive Bayes, and neural network algorithms. According to the GWAS, rs2110179, rs7126100, and rs2076549 were associated with the development of peripheral neuropathy on vincristine administration. Machine learning was performed using these three SNPs to construct a prediction model. A high accuracy of 93.8% was obtained with the support vector machine and neural network using rs2110179 and rs2076549. Thus, peripheral neuropathy development due to vincristine therapy can be effectively predicted by a machine learning prediction model using SNPs associated with it.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Pharmacogenomics Journal
医学-药学
CiteScore
7.20
自引率
0.00%
发文量
35
审稿时长
6-12 weeks
期刊介绍:
The Pharmacogenomics Journal is a print and electronic journal, which is dedicated to the rapid publication of original research on pharmacogenomics and its clinical applications.
Key areas of coverage include:
Personalized medicine
Effects of genetic variability on drug toxicity and efficacy
Identification and functional characterization of polymorphisms relevant to drug action
Pharmacodynamic and pharmacokinetic variations and drug efficacy
Integration of new developments in the genome project and proteomics into clinical medicine, pharmacology, and therapeutics
Clinical applications of genomic science
Identification of novel genomic targets for drug development
Potential benefits of pharmacogenomics.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


