新生小鼠中Nf1、Pten和Trp53的基因组编辑可诱导少突胶质细胞谱系转录因子2阳性的胶质母细胞瘤。
IF 0.9
4区 医学
Q4 PATHOLOGY
Journal of Toxicologic Pathology
Pub Date : 2021-10-01
Epub Date: 2021-07-23
DOI:10.1293/tox.2021-0029
引用次数: 1
摘要
为了通过基因组编辑生成小鼠胶质母细胞瘤模型,我们通过电穿孔将Cas9蛋白和Nf1、Pten和Trp53特异性的引导rna引入新生小鼠前脑。我们发现,15周后,胶质肿瘤(包括胶质母细胞瘤)的发生率很高(约90%)。肿瘤的组织学特征与弥漫性胶质瘤相似,在某些情况下,与人类胶质母细胞瘤相似,微血管增生(肾小球结构)。此外,与先前模型中使用类似方法生成的胶质原纤维酸性蛋白(GFAP)阳性胶质母细胞瘤不同,大多数肿瘤细胞对少突胶质细胞谱系转录因子2呈阳性,但对GFAP和神经丝呈阴性。在Nf1, Pten和Trp53的靶序列周围发现了一个与先前模型中相同的碱基对插入,并且仅在Pten中发现了额外的缺失。考虑到组织学特征与之前模型不同,我们的新模型为研究胶质母细胞瘤的早期发展提供了额外的研究工具。本文章由计算机程序翻译,如有差异,请以英文原文为准。
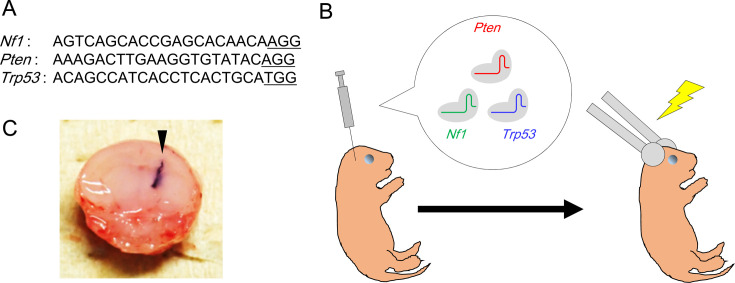
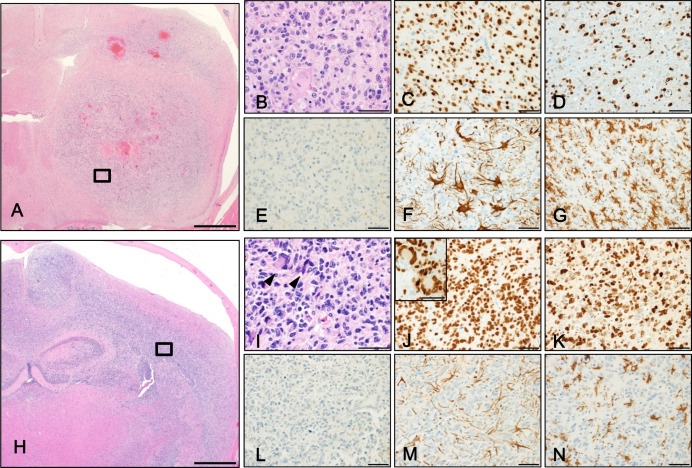
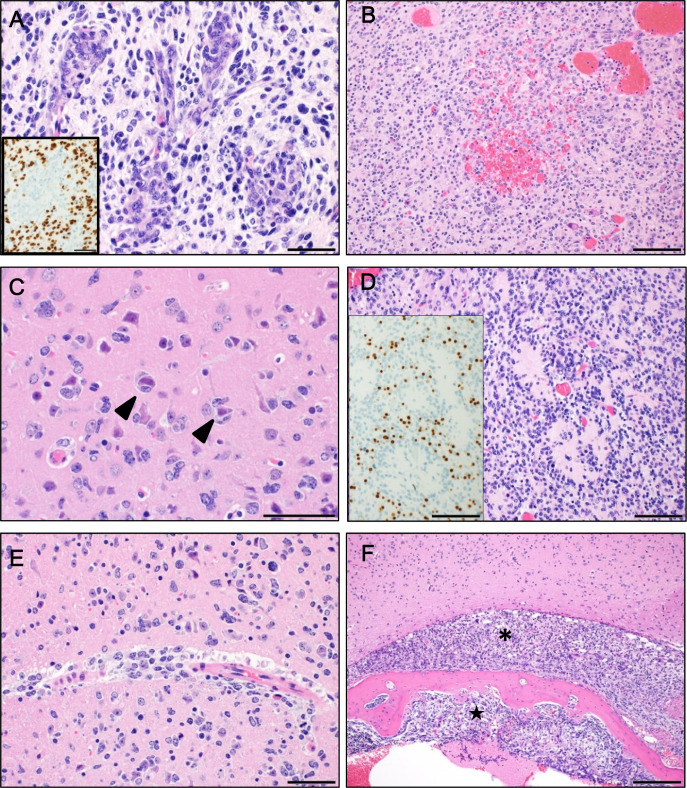
Genome editing of Nf1, Pten, and Trp53 in neonatal mice induces glioblastomas positive for oligodendrocyte lineage transcription factor 2.
To generate a mouse glioblastoma model by genome editing, we introduced Cas9 protein and guide RNAs specific for Nf1, Pten, and Trp53 into the neonatal mouse forebrain by electroporation. We found a high incidence (approximately 90%) of glial tumor development, including glioblastomas, 15 weeks later. The histological features of the tumors were similar to those of diffuse gliomas and, in some cases, similar to human glioblastomas, with microvascular proliferation (glomeruloid structure). In addition, unlike glial fibrillary acidic protein (GFAP)-positive glioblastomas generated using a similar method in a previous model, the majority of tumor cells were positive for oligodendrocyte lineage transcription factor 2, but negative for GFAP and neurofilaments. One base pair insertions identical to those seen in a previous model were found around the target sequences in Nf1, Pten, and Trp53, and additional deletions were found only in Pten. Considering that the histological characteristics were different from those seen in the previous model, our new model provides an additional research tool to investigate the early stages of glioblastoma development.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Toxicologic Pathology
PATHOLOGY-TOXICOLOGY
CiteScore
2.10
自引率
16.70%
发文量
22
审稿时长
>12 weeks
期刊介绍:
JTP is a scientific journal that publishes original studies in the field of toxicological pathology and in a wide variety of other related fields. The main scope of the journal is listed below.
Administrative Opinions of Policymakers and Regulatory Agencies
Adverse Events
Carcinogenesis
Data of A Predominantly Negative Nature
Drug-Induced Hematologic Toxicity
Embryological Pathology
High Throughput Pathology
Historical Data of Experimental Animals
Immunohistochemical Analysis
Molecular Pathology
Nomenclature of Lesions
Non-mammal Toxicity Study
Result or Lesion Induced by Chemicals of Which Names Hidden on Account of the Authors
Technology and Methodology Related to Toxicological Pathology
Tumor Pathology; Neoplasia and Hyperplasia
Ultrastructural Analysis
Use of Animal Models.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


