Suzanne E. Dettling, Mina Ahmadi, Zongtao Lin, Lin He, Megan L. Matthews
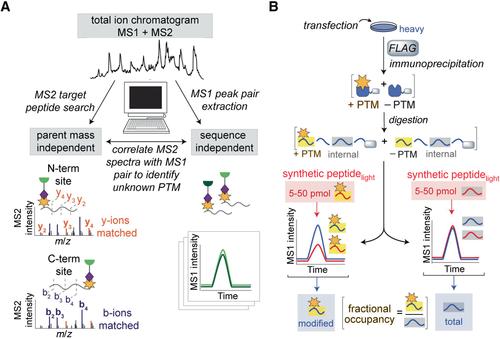
{"title":"亲电试剂的发现和酶辅助因子的分析","authors":"Suzanne E. Dettling, Mina Ahmadi, Zongtao Lin, Lin He, Megan L. Matthews","doi":"10.1002/cpch.86","DOIUrl":null,"url":null,"abstract":"<p>Reverse-polarity activity-based protein profiling (RP-ABPP) is a chemical proteomics approach that uses nucleophilic probes amenable to “click” chemistry deployed into living cells in culture to capture, immunoprecipitate, and identify protein-bound electrophiles. RP-ABPP is used to characterize the structure and function of reactive electrophilic post-translational modifications (PTMs) and the proteins harboring them, which may uncover unknown or novel functions. RP-ABPP has demonstrated utility as a versatile method to monitor the metabolic regulation of electrophilic cofactors, using a pyruvoyl cofactor in <i>S</i>-adenosyl-<span>L</span>-methionine decarboxylase (AMD1), and to discover novel types of electrophilic modifications on proteins in human cells, such as the glyoxylyl modification on secernin-3 (SCRN3). These cofactors cannot be predicted by sequence, and therefore this area is relatively undeveloped. RP-ABPP is the only global, unbiased approach to discover such electrophiles. Here, we describe the utility of these experiments and provide a detailed protocol for de novo discovery, quantitation, and global profiling of electrophilic functionality of proteins. © 2020 The Authors.</p><p><b>Basic Protocol 1</b>: Identification and quantification of probe-reactive proteins</p><p><b>Basic Protocol 2</b>: Characterization of the site of probe labeling</p><p><b>Basic Protocol 3</b>: Determination and quantitation of electrophile structure</p>","PeriodicalId":38051,"journal":{"name":"Current protocols in chemical biology","volume":"12 4","pages":""},"PeriodicalIF":0.0000,"publicationDate":"2020-11-16","publicationTypes":"Journal Article","fieldsOfStudy":null,"isOpenAccess":false,"openAccessPdf":"https://sci-hub-pdf.com/10.1002/cpch.86","citationCount":"8","resultStr":"{\"title\":\"Discovery of Electrophiles and Profiling of Enzyme Cofactors\",\"authors\":\"Suzanne E. Dettling, Mina Ahmadi, Zongtao Lin, Lin He, Megan L. Matthews\",\"doi\":\"10.1002/cpch.86\",\"DOIUrl\":null,\"url\":null,\"abstract\":\"<p>Reverse-polarity activity-based protein profiling (RP-ABPP) is a chemical proteomics approach that uses nucleophilic probes amenable to “click” chemistry deployed into living cells in culture to capture, immunoprecipitate, and identify protein-bound electrophiles. RP-ABPP is used to characterize the structure and function of reactive electrophilic post-translational modifications (PTMs) and the proteins harboring them, which may uncover unknown or novel functions. RP-ABPP has demonstrated utility as a versatile method to monitor the metabolic regulation of electrophilic cofactors, using a pyruvoyl cofactor in <i>S</i>-adenosyl-<span>L</span>-methionine decarboxylase (AMD1), and to discover novel types of electrophilic modifications on proteins in human cells, such as the glyoxylyl modification on secernin-3 (SCRN3). These cofactors cannot be predicted by sequence, and therefore this area is relatively undeveloped. RP-ABPP is the only global, unbiased approach to discover such electrophiles. Here, we describe the utility of these experiments and provide a detailed protocol for de novo discovery, quantitation, and global profiling of electrophilic functionality of proteins. © 2020 The Authors.</p><p><b>Basic Protocol 1</b>: Identification and quantification of probe-reactive proteins</p><p><b>Basic Protocol 2</b>: Characterization of the site of probe labeling</p><p><b>Basic Protocol 3</b>: Determination and quantitation of electrophile structure</p>\",\"PeriodicalId\":38051,\"journal\":{\"name\":\"Current protocols in chemical biology\",\"volume\":\"12 4\",\"pages\":\"\"},\"PeriodicalIF\":0.0000,\"publicationDate\":\"2020-11-16\",\"publicationTypes\":\"Journal Article\",\"fieldsOfStudy\":null,\"isOpenAccess\":false,\"openAccessPdf\":\"https://sci-hub-pdf.com/10.1002/cpch.86\",\"citationCount\":\"8\",\"resultStr\":null,\"platform\":\"Semanticscholar\",\"paperid\":null,\"PeriodicalName\":\"Current protocols in chemical biology\",\"FirstCategoryId\":\"1085\",\"ListUrlMain\":\"https://onlinelibrary.wiley.com/doi/10.1002/cpch.86\",\"RegionNum\":0,\"RegionCategory\":null,\"ArticlePicture\":[],\"TitleCN\":null,\"AbstractTextCN\":null,\"PMCID\":null,\"EPubDate\":\"\",\"PubModel\":\"\",\"JCR\":\"Q3\",\"JCRName\":\"Biochemistry, Genetics and Molecular Biology\",\"Score\":null,\"Total\":0}","platform":"Semanticscholar","paperid":null,"PeriodicalName":"Current protocols in chemical biology","FirstCategoryId":"1085","ListUrlMain":"https://onlinelibrary.wiley.com/doi/10.1002/cpch.86","RegionNum":0,"RegionCategory":null,"ArticlePicture":[],"TitleCN":null,"AbstractTextCN":null,"PMCID":null,"EPubDate":"","PubModel":"","JCR":"Q3","JCRName":"Biochemistry, Genetics and Molecular Biology","Score":null,"Total":0}
引用次数: 8

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


