基于种子的 DNA 序列最佳压缩算法
Q1 Biochemistry, Genetics and Molecular Biology
引用次数: 0
摘要
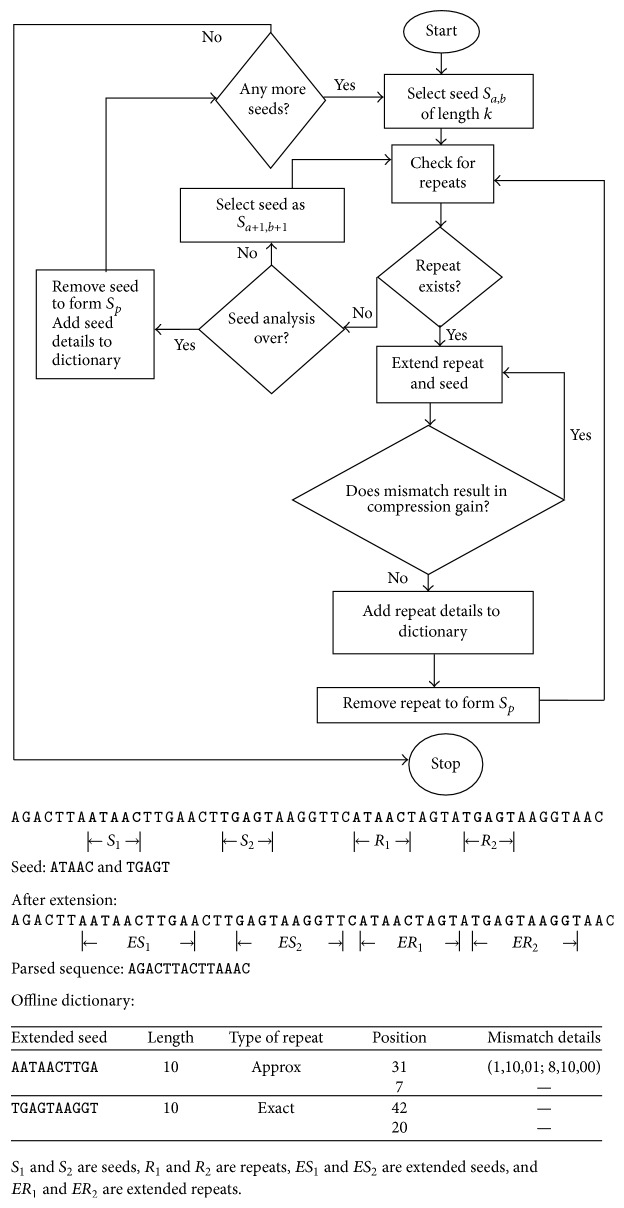
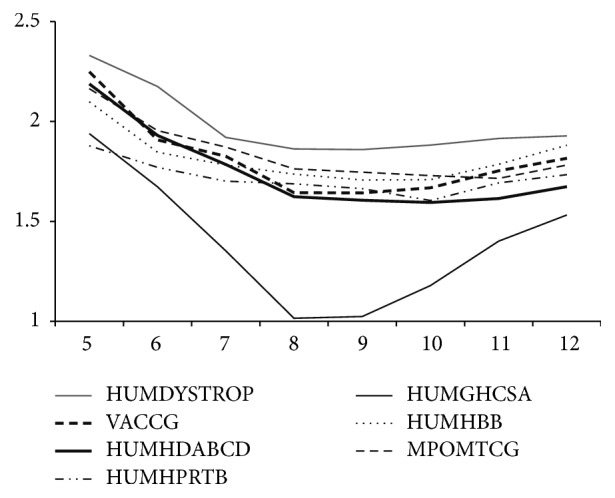
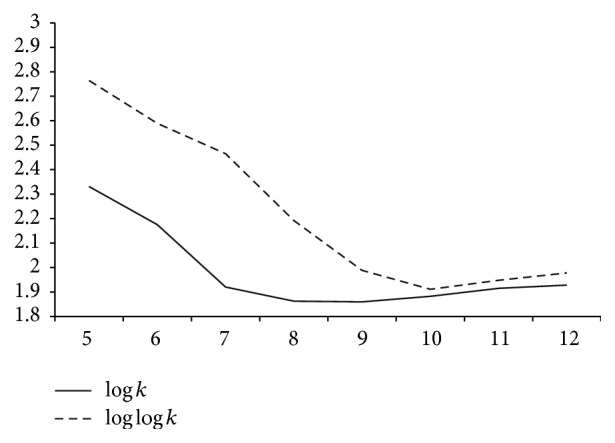
本文提出了一种基于种子的无损压缩算法来压缩 DNA 序列,该算法使用的替换方法与 LempelZiv 压缩方案类似。所提出的方法利用了 DNA 序列中固有的重复结构,创建了一个离线字典,其中包含所有此类重复以及错配的详细信息。通过确保只允许有希望的错配,该方法实现了与现有无损 DNA 序列压缩算法相当或更好的压缩率。本文章由计算机程序翻译,如有差异,请以英文原文为准。
An Optimal Seed Based Compression Algorithm for DNA Sequences.
This paper proposes a seed based lossless compression algorithm to compress a DNA sequence which uses a substitution method that is similar to the LempelZiv compression scheme. The proposed method exploits the repetition structures that are inherent in DNA sequences by creating an offline dictionary which contains all such repeats along with the details of mismatches. By ensuring that only promising mismatches are allowed, the method achieves a compression ratio that is at par or better than the existing lossless DNA sequence compression algorithms.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Advances in Bioinformatics
Biochemistry, Genetics and Molecular Biology-Biochemistry, Genetics and Molecular Biology (miscellaneous)
自引率
0.00%
发文量
0
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


