通过纳米孔测序确定复杂mrna的外显子连通性。
IF 12.3
1区 生物学
Q1 Agricultural and Biological Sciences
引用次数: 134
摘要
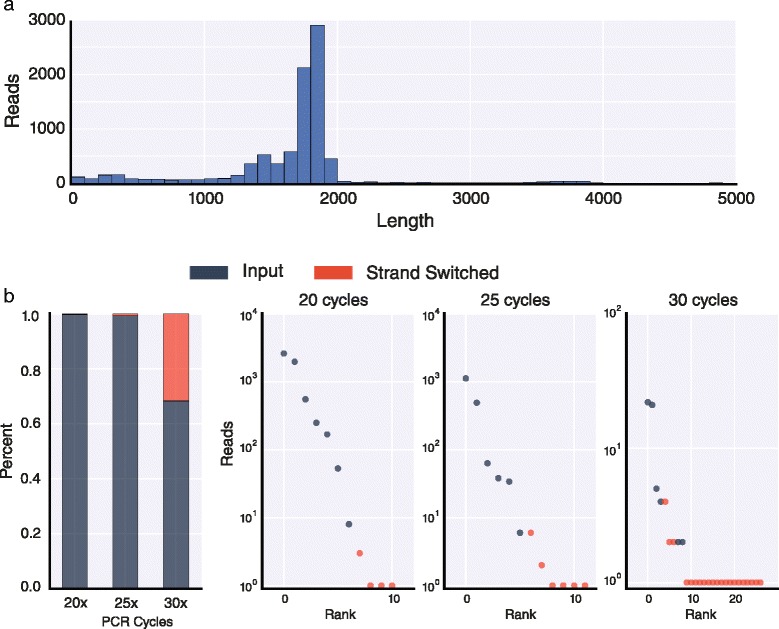
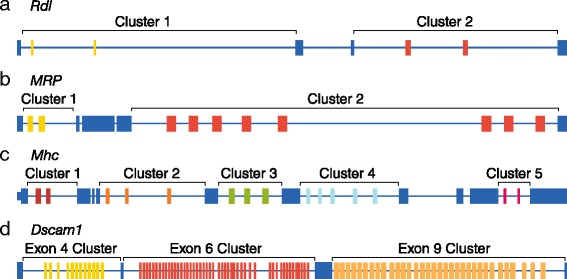
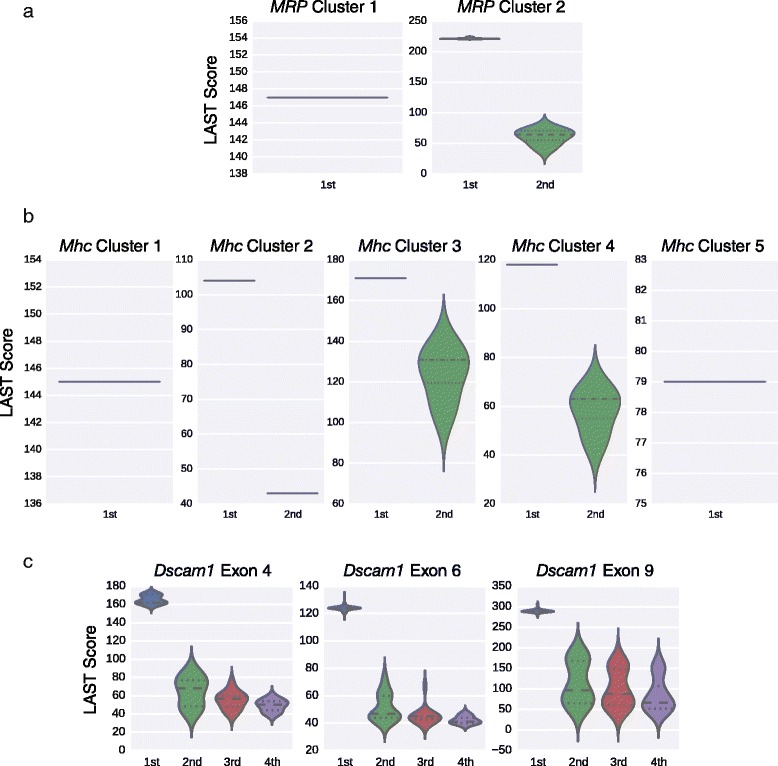
短读高通量RNA测序虽然功能强大,但其直接测量mrna外显子连通性的能力有限,这些mrna包含多个可选外显子,这些外显子位于比最大阅读长度更远的地方。在这里,我们使用牛津纳米孔MinION测序仪鉴定了7899个“全长”同种异构体,表达自果蝇的四个基因,Dscam1, MRP, Mhc和Rdl。这些结果表明,纳米孔测序可以用于解卷积单个亚型,并且它有潜力成为全面转录组表征的有力方法。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Determining exon connectivity in complex mRNAs by nanopore sequencing.
Short-read high-throughput RNA sequencing, though powerful, is limited in its ability to directly measure exon connectivity in mRNAs that contain multiple alternative exons located farther apart than the maximum read length. Here, we use the Oxford Nanopore MinION sequencer to identify 7,899 'full-length' isoforms expressed from four Drosophila genes, Dscam1, MRP, Mhc, and Rdl. These results demonstrate that nanopore sequencing can be used to deconvolute individual isoforms and that it has the potential to be a powerful method for comprehensive transcriptome characterization.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Genome Biology
BIOTECHNOLOGY & APPLIED MICROBIOLOGY-GENETICS & HEREDITY
CiteScore
25.50
自引率
3.30%
发文量
0
审稿时长
14 weeks
期刊介绍:
Genome Biology is a leading research journal that focuses on the study of biology and biomedicine from a genomic and post-genomic standpoint. The journal consistently publishes outstanding research across various areas within these fields.
With an impressive impact factor of 12.3 (2022), Genome Biology has earned its place as the 3rd highest-ranked research journal in the Genetics and Heredity category, according to Thomson Reuters. Additionally, it is ranked 2nd among research journals in the Biotechnology and Applied Microbiology category. It is important to note that Genome Biology is the top-ranking open access journal in this category.
In summary, Genome Biology sets a high standard for scientific publications in the field, showcasing cutting-edge research and earning recognition among its peers.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


