基于蛋白质-蛋白质对接模拟和参数调优的亲和性评估与预测系统的开发。
Q2 Biochemistry, Genetics and Molecular Biology
Advances and Applications in Bioinformatics and Chemistry
Pub Date : 2009-01-01
Epub Date: 2009-01-12
DOI:10.2147/aabc.s3646
引用次数: 19
摘要
利用广泛应用的形状互补搜索方法作为蛋白质对接仿真算法,建立了蛋白质对相互作用的评估和预测系统。我们使用这个系统,我们称之为亲和力评价和预测(AEP)系统,评估了20对蛋白质之间的相互作用。该系统首先对目标蛋白群进行“轮循式”形状互补搜索,并评估搜索得到的复杂结构之间的相互作用。这些复杂的结构是通过使用我们开发的称为“分组”的统计程序来选择的。在5.0%的流行率下,我们的AEP系统预测蛋白质-蛋白质相互作用的召回率为50.0%,精密度为55.6%,准确度为95.5%,f值为0.526。通过优化分组过程,我们的AEP系统成功预测了20对蛋白对中的10对具有生物学相关性的组合。我们的最终目标是建立一个亲和数据库,为细胞生物学家和药物设计者提供使用我们的AEP系统获得的关键信息。本文章由计算机程序翻译,如有差异,请以英文原文为准。
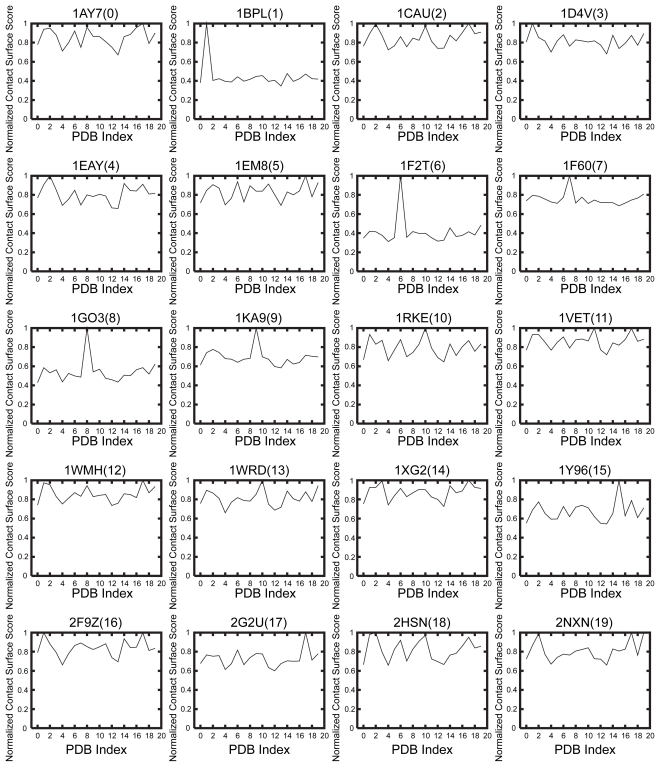
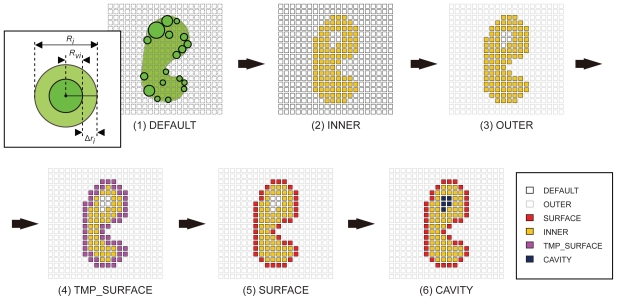
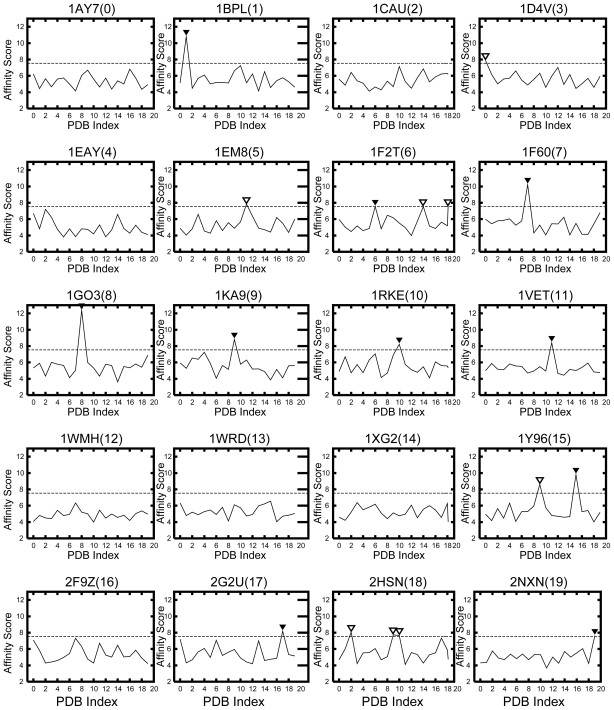
The development of an affinity evaluation and prediction system by using protein-protein docking simulations and parameter tuning.
A system was developed to evaluate and predict the interaction between protein pairs by using the widely used shape complementarity search method as the algorithm for docking simulations between the proteins. We used this system, which we call the affinity evaluation and prediction (AEP) system, to evaluate the interaction between 20 protein pairs. The system first executes a “round robin” shape complementarity search of the target protein group, and evaluates the interaction between the complex structures obtained by the search. These complex structures are selected by using a statistical procedure that we developed called ‘grouping’. At a prevalence of 5.0%, our AEP system predicted protein–protein interactions with a 50.0% recall, 55.6% precision, 95.5% accuracy, and an F-measure of 0.526. By optimizing the grouping process, our AEP system successfully predicted 10 protein pairs (among 20 pairs) that were biologically relevant combinations. Our ultimate goal is to construct an affinity database that will provide cell biologists and drug designers with crucial information obtained using our AEP system.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Advances and Applications in Bioinformatics and Chemistry
Biochemistry, Genetics and Molecular Biology-Biochemistry, Genetics and Molecular Biology (miscellaneous)
CiteScore
6.50
自引率
0.00%
发文量
7
审稿时长
16 weeks
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


