一种结合基于结构的药物筛选和基于配体的筛选来提高命中率的方法。
Q2 Biochemistry, Genetics and Molecular Biology
Advances and Applications in Bioinformatics and Chemistry
Pub Date : 2008-01-01
Epub Date: 2008-08-12
DOI:10.2147/aabc.s3767
引用次数: 11
摘要
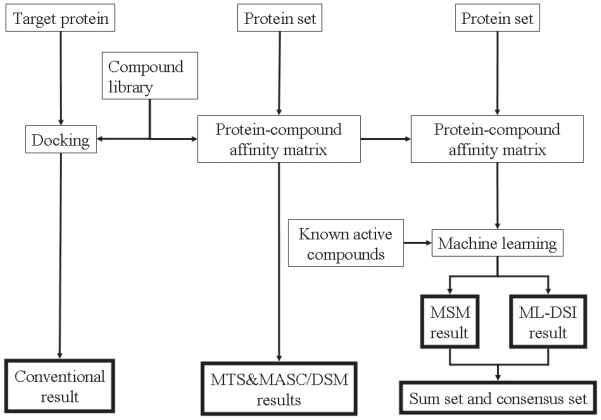
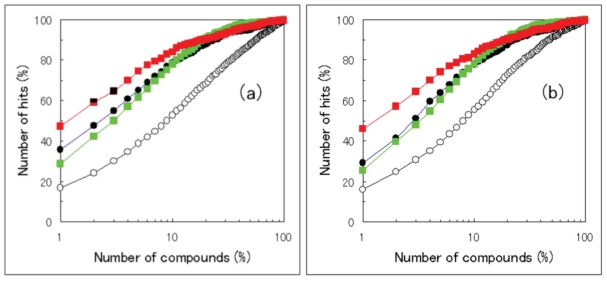
我们研究了将两种不同的硅药物筛选结果结合起来的程序,以达到高命中率。当已知目标蛋白和一些活性化合物的三维结构时,基于结构和基于配体的硅基筛选方法都可以应用。本研究采用机器学习评分修改多靶点筛选(MSM-MTS)方法作为基于结构的筛选方法,采用机器学习对接评分指数(ML-DSI)方法作为基于配体的筛选方法。为了结合这两种筛选方法的预测化合物集,我们检查了集合的乘积(共识集)和集合的和。结果,共识集的命中率高于集合的总和,也高于任何一个单独的预测集。此外,在分子动力学模拟中,目前的组合对于不同晶体结构和快照结构的结构多样性都显示出足够的鲁棒性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
A method to enhance the hit ratio by a combination of structure-based drug screening and ligand-based screening.
We examined the procedures to combine two different in silico drug-screening results to achieve a high hit ratio. When the 3D structure of the target protein and some active compounds are known, both structure-based and ligand-based in silico screening methods can be applied. In the present study, the machine-learning score modification multiple target screening (MSM-MTS) method was adopted as a structure-based screening method, and the machine-learning docking score index (ML-DSI) method was adopted as a ligand-based screening method. To combine the predicted compound’s sets by these two screening methods, we examined the product of the sets (consensus set) and the sum of the sets. As a result, the consensus set achieved a higher hit ratio than the sum of the sets and than either individual predicted set. In addition, the current combination was shown to be robust enough for the structural diversities both in different crystal structure and in snapshot structures during molecular dynamics simulations.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Advances and Applications in Bioinformatics and Chemistry
Biochemistry, Genetics and Molecular Biology-Biochemistry, Genetics and Molecular Biology (miscellaneous)
CiteScore
6.50
自引率
0.00%
发文量
7
审稿时长
16 weeks
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


