基于仿真的心电图激活和复极序列的数字孪生横跨健康和患病心脏。
IF 6.3
2区 医学
Q1 BIOLOGY
引用次数: 0
摘要
心室复极异常模式被认为是导致各种心脏疾病致死性心律失常的原因,包括遗传性和获得性通道病、心肌病和缺血性心脏病。然而,检测这些重极化异常的方法是有限的。在这项研究中,我们介绍并评估了一种基于模拟的新方法,该方法可以从12导联心电图(ECG)和磁共振衍生的心室解剖重建中推断心室激活和复极时间,首次适用于健康对照和复极异常病例。首先,通过对早期激活位点和传导速度的迭代细化,重建心室激活时间,直到模型和目标QRS复合物匹配。然后,通过反复细化心室动作电位持续时间和动作电位形状参数来重建心室复极时间,直到模型和目标T波匹配,包括正则化。根据已知复极化时间的18个基准模拟评估复极化推断,包括对照组和肥厚性心肌病(HCM)异常复极化病例。推断的复极化时间与对照和HCM条件下的真实情况吻合较好(Spearman r分别为0.63±0.11和0.65±0.19),推断的模型T波与目标T波吻合较好(r分别为0.81±0.05和0.78±0.08)。该方法在重建一系列异常心室复极序列的延迟复极宏观模式方面显示出灵活性,证明了对广泛病理情景的适用性。基于模拟的推断可以准确地从12导联心电图中重建正常和异常复极模式的复极时间。本文章由计算机程序翻译,如有差异,请以英文原文为准。
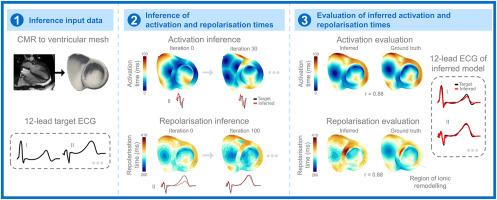
Simulation-based digital twinning of activation and repolarisation sequences from the ECG across healthy and diseased hearts
Abnormal patterns of ventricular repolarisation are thought to contribute to lethal arrhythmias in various cardiac conditions, including inherited and acquired channelopathies, cardiomyopathies, and ischaemic heart disease. However, methods to detect these repolarisation abnormalities are limited.
In this study, we introduce and assess a novel simulation-based method to infer ventricular activation and repolarisation times from the 12-lead electrocardiogram (ECG) and magnetic resonance-derived ventricular anatomical reconstruction, applicable for the first time to both healthy controls and cases with abnormal repolarisation.
First, ventricular activation times were reconstructed through iterative refinement of early activation sites and conduction velocities, until the model and target QRS complexes matched. Then, ventricular repolarisation times were reconstructed through iterative refinement of ventricular action potential durations and an action potential shape parameter until the model and target T waves matched, including regularisation. Repolarisation inference was evaluated against 18 benchmark simulations with known repolarisation times, including both control and hypertrophic cardiomyopathy (HCM) cases with abnormal repolarisation.
Inferred repolarisation times showed good agreement with the ground truth in control and HCM (Spearman and , respectively), with the inferred model T waves closely matching the target T waves ( and , respectively). The method further demonstrated flexibility in reconstructing the macroscopic patterns of delayed repolarisation across a range of abnormal ventricular repolarisation sequences, demonstrating applicability to a wide range of pathological scenarios.
Simulation-based inference can accurately reconstruct repolarisation times from the 12-lead ECG in cases with both normal and abnormal repolarisation patterns.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


