基于深度学习的CECT图像自动检测化脓性肝脓肿和肺炎克雷伯菌感染的诊断:一项多中心研究
IF 3.3
3区 医学
Q1 RADIOLOGY, NUCLEAR MEDICINE & MEDICAL IMAGING
引用次数: 0
摘要
在低收入和中等收入国家,准确鉴定化脓性肝脓肿(PLA)的细菌病原体仍然具有挑战性。我们开发并验证了一种人工智能(AI)模型,该模型可以复制临床工作流程,使用对比增强计算机断层扫描(CECT)检测和诊断肺炎克雷伯菌肝脓肿(KPLA)。方法于2011年4月至2024年4月从3个医疗中心招募PLA患者。人工智能化脓性肝脓肿检测(PLADA)系统采用V-Net深度神经网络进行病灶自动分割。随后提取放射组学特征并按重要性排序。六种机器学习算法用于开发临床、放射组学和综合临床-影像学模型,用于培训队列中的KPLA诊断,并在多中心框架中进行外部验证。结果共发现492例PLA患者。PLADA算法准确分割病灶,平均骰子系数为0.941(95%可信区间:0.899-0.983)。临床成像模型表现出很强的歧视性,在训练队列中所有算法的平均曲线下面积(AUC)超过0.9。基于adaboost的临床成像模型在外部验证队列中的AUC为0.847 (95% CI: 0.753-0.940),具有高灵敏度(71.9%)和特异性(86.4%)。决策曲线分析(DCA)证实了临床应用。结论PLADA系统通过临床可解释的两阶段人工智能框架,为KPLA诊断提供了一种准确、无创的方法。虽然受限于回顾性的单一国家数据集,但这种方法在资源有限的情况下对KPLA的早期检测显示出特别的希望。本文章由计算机程序翻译,如有差异,请以英文原文为准。
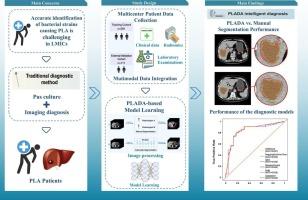
Automated detection of pyogenic liver abscess and diagnosis of Klebsiella pneumoniae infection based on CECT images with deep learning: A multicenter study
Background
Accurate identification of bacterial pathogens in pyogenic liver abscess (PLA) remains challenging in low- and middle-income countries (LMICs). We developed and validated an artificial intelligence (AI) model that replicates clinical workflow to detect and diagnose Klebsiella pneumoniae liver abscess (KPLA) using contrast-enhanced computerized tomography (CECT).
Methods
Patients with PLA were enrolled from three medical centers between April 2011 and April 2024. The Pyogenic Liver Abscess Detection with AI (PLADA) system employed a V-Net deep neural network for automated lesion segmentation. Radiomics features were subsequently extracted and ranked by importance. Six machine learning algorithms were used to develop clinical, radiomics, and integrated clinical-imaging models for KPLA diagnosis in the training cohort, with external validation in a multicenter framework.
Results
A total of 492 PLA patients were identified. The PLADA algorithm accurately segments the lesions with a mean dice coefficient of 0.941 (95 % confidence interval: 0.899–0.983). The clinical-imaging model demonstrated strong discriminatory performance, with a mean area under the curve (AUC) of over 0.9 across all algorithms in the training cohort. The AdaBoost-based clinical-imaging model achieved an AUC of 0.847 (95 % CI: 0.753–0.940) in the external validation cohort, with high sensitivity (71.9 %) and specificity (86.4 %). Decision curve analysis (DCA) confirmed clinical utility.
Conclusions
The PLADA system provides an accurate, non-invasive method for KPLA diagnosis through a clinically interpretable two-stage AI framework. While limited by a retrospective, single-country dataset, this approach shows particular promise for early detection of KPLA in resource-limited settings.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
6.70
自引率
3.00%
发文量
398
审稿时长
42 days
期刊介绍:
European Journal of Radiology is an international journal which aims to communicate to its readers, state-of-the-art information on imaging developments in the form of high quality original research articles and timely reviews on current developments in the field.
Its audience includes clinicians at all levels of training including radiology trainees, newly qualified imaging specialists and the experienced radiologist. Its aim is to inform efficient, appropriate and evidence-based imaging practice to the benefit of patients worldwide.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


