可解释支持向量分类器可靠预测修饰肽对大肠杆菌的抗菌活性
IF 3
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
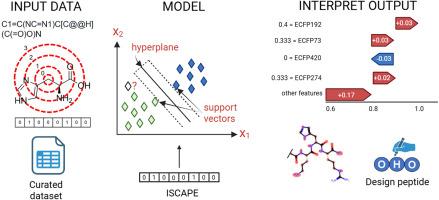
抗菌肽(AMPs)是传统抗生素的有希望的替代品,传统抗生素的有效性由于抗菌素耐药性(AMR)的上升而下降。为了加速AMP的发现,我们开发了ISCAPE(肽抗大肠杆菌抗菌活性的可解释支持向量分类器),这是一种机器学习(ML)模型,解决了当前AMP预测器的局限性。ISCAPE只需要一个简化的分子输入行输入系统(SMILES)字符串作为输入,就可以预测天然和化学修饰肽对大肠杆菌ATCC 25922的活性。最低抑制浓度(MIC)阈值≤16 μg/mL。为了确保可靠性,只有在可比较的实验条件下获得的MIC值被包括在我们整理的数据集中。ISCAPE优于最先进的antipmod,接收器工作特性曲线下面积(AUROC)为91.83%,马修相关系数(MCC)为71.86%。驱动这种性能的特征包括碳-碳对的比例以及基于特征和计数的扩展连接指纹(ecfp)。通过SHapley加性解释(SHAP)增强了模型的可解释性,该解释确定了AMP活性最关键的分子特征。据我们所知,ISCAPE是第一个可解释的ML预测器,能够预测天然和修饰肽对特定大肠杆菌菌株的抗菌活性。它是一种用户友好的工具,允许实验人员确定关键的分子特征,减少对广泛的结构-活性关系(SAR)研究的需要,并指导新型amp的设计。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Interpretable support vector classifier for reliable prediction of antibacterial activity of modified peptides against Escherichia coli
Antimicrobial peptides (AMPs) are promising alternatives to traditional antibiotics, whose effectiveness is declining due to rising antimicrobial resistance (AMR). To accelerate AMP discovery, we developed ISCAPE (Interpretable Support Vector Classifier of Antibacterial Activity of Peptides against Escherichia coli), a machine learning (ML) model that addresses the limitations of current AMP predictors. ISCAPE requires only a Simplified Molecular-Input Line-Entry System (SMILES) string as input and can predict the activity of both natural and chemically modified peptides against E. coli ATCC 25922. Activity is defined by a minimum inhibitory concentration (MIC) threshold of ≤16 μg/mL. To ensure reliability, only MIC values obtained under comparable experimental conditions were included in our curated dataset. ISCAPE outperformed the state-of-the-art AntiMPmod, achieving an area under the receiver operating characteristic curve (AUROC) of 91.83% and a Matthew's correlation coefficient (MCC) of 71.86%. Features driving this performance include the fraction of carbon-carbon pairs and feature- and count-based extended connectivity fingerprints (ECFPs). Model interpretability is enhanced through SHapley Additive exPlanations (SHAP), which identifies the molecular features most critical for AMP activity. To our knowledge, ISCAPE is the first interpretable ML predictor capable of predicting antibacterial activity for both natural and modified peptides against a specific E. coli strain. It is a user-friendly tool that allows experimentalists to pinpoint key molecular features, reducing the need for extensive structure-activity relationship (SAR) studies and guiding the design of novel AMPs.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


