一个REDCap高级随机化模块,以满足现代试验的需要。
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
目的:自2012年以来,电子数据采集平台REDCap包含了一个嵌入式随机化模块,允许每个研究记录进行单个随机化,并能够根据研究地点和参与者出生时的性别等变量进行分层。近年来,平台化、自适应、去中心化、实用化的审判越来越受欢迎。这些试验设计通常需要采用原始REDCap随机化模块不支持的随机化方法,包括将患者随机分配到多个领域或多个时间点,改变分配表以增加或减少研究组,或根据先前入组的参与者的数据自适应地改变分配比例。我们的团队旨在开发新的随机化功能来解决这些问题。方法:在美国国立卫生研究院资助的试验创新网络的推动下,启动了一个协作过程,将临床试验学家、生物统计学家、技术专家和其他专家的反馈结合起来,使REDCap中的随机化模块现代化。结果:这一努力促成了REDCap平台内高级随机化模块的开发。除了支持平台、自适应、去中心化和实用的试验之外,新模块还引入了几个新功能,例如改进了对盲法随机化的支持、额外的随机化元数据捕获(例如,用户身份和时间戳)、允许REDCap管理员使用随机化模块支持调查人员的额外工具。参与实用或分散试验的临床医生无需登录研究数据库即可通过调查执行随机化的能力。截至2025年6月19日,55所院校211个项目采用了多重随机化,64所院校108个项目采用了实时触发逻辑随机化,17所院校24个项目采用了盲法分组。结论:新的随机化模块旨在简化随机化过程,提高试验效率,确保数据的完整性,从而支持开展更复杂和适应性更强的临床试验。本文章由计算机程序翻译,如有差异,请以英文原文为准。
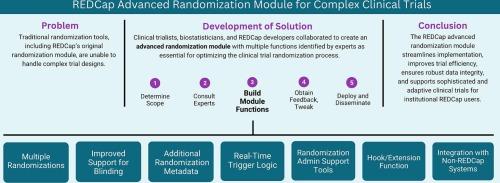
A REDCap advanced randomization module to meet the needs of modern trials
Objective
Since 2012, the electronic data capture platform REDCap has included an embedded randomization module allowing a single randomization per study record with the ability to stratify by variables such as study site and participant sex at birth. In recent years, platform, adaptive, decentralized, and pragmatic trials have gained popularity. These trial designs often require approaches to randomization not supported by the original REDCap randomization module, including randomizing patients into multiple domains or at multiple points in time, changing allocation tables to add or drop study groups, or adaptively changing allocation ratios based on data from previously enrolled participants. Our team aimed to develop new randomization functions to address these issues.
Methods
A collaborative process facilitated by the NIH-funded Trial Innovation Network was initiated to modernize the randomization module in REDCap, incorporating feedback from clinical trialists, biostatisticians, technologists, and other experts.
Results
This effort led to the development of an advanced randomization module within the REDCap platform. In addition to supporting platform, adaptive, decentralized, and pragmatic trials, the new module introduces several new features, such as improved support for blinded randomization, additional randomization metadata capture (e.g., user identity and timestamp), additional tools allowing REDCap administrators to support investigators using the randomization module, and the ability for clinicians participating in pragmatic or decentralized trials to perform randomization through a survey without needing log-in access to the study database. As of June 19, 2025, multiple randomizations have been used in 211 projects from 55 institutions, randomizations with real-time trigger logic in 108 projects from 64 institutions, and blinded group allocation in 24 projects from 17 institutions.
Conclusion
The new randomization module aims to streamline the randomization process, improve trial efficiency, and ensure robust data integrity, thereby supporting the conduct of more sophisticated and adaptive clinical trials.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


