MBT-Polyp:一种新的用于息肉分割的多分支内存增强变压器
IF 4.2
3区 计算机科学
Q2 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
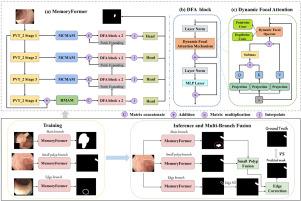
息肉分割对结直肠癌的早期诊断和临床精准干预具有重要意义。尽管深度学习在医学图像分割方面取得了重大进展,但在结肠直肠息肉分割中,小息肉的准确定位和息肉边界的精确划定仍然是一个挑战。在这项研究中,我们引入了MBT-Polyp,一种多分支内存增强变压器架构,旨在提高对小息肉的分割灵敏度,并提高对模糊息肉边界的描绘精度。我们的框架的核心是MemoryFormer,这是一个基于transformer的u形架构,包含三个关键组件:用于高效小目标增强和边缘细化的动态焦点注意块(DFA),通过交叉分辨率融合保存边界细节的高级内存注意模块(hmm),以及用于抑制背景噪声和建模局部空间上下文的多视图通道内存注意模块(MCMAM)。为了指导专业学习,我们在ground truth旁边导出小息肉和边缘标签,使MemoryFormer能够通过专用分支处理它们。输出使用小息肉融合策略(SPFS)和边缘校正策略(ECS)进行融合,以减轻过度和欠分割。Kvasir-SEG、CVC-ColonDB、CVC-ClinicDB、CVC-300和ETIS-Larib的定量结果平均Dice评分分别为0.930、0.818、0.943、0.912和0.763,在不同的息肉分割场景中具有很强的泛化性。代码和数据集可在:https://github.com/taojlu/PolypSeg。本文章由计算机程序翻译,如有差异,请以英文原文为准。
MBT-Polyp: A new Multi-Branch Memory-augmented Transformer for polyp segmentation
Polyp segmentation plays a critical role in the early diagnosis and precise clinical intervention of colorectal cancer (CRC). Despite significant advancements in deep learning for medical image segmentation, accurate localization of small polyps and precise delineation of polyp boundaries remain challenges in colorectal polyp segmentation. In this study, we introduce MBT-Polyp, a Multi-branch Memory-augmented Transformer architecture designed to improve segmentation sensitivity for small polyps and enhance the delineation accuracy of ambiguous polyp boundaries. At the core of our framework is MemoryFormer, a Transformer-based U-shaped architecture that incorporates three key components: a Dynamic Focal Attention block (DFA) for efficient small target enhancement and edge refinement, a High-Level Memory Attention Module (HMAM) for preserving boundary details via cross-resolution fusion, and a Multi-View Channel Memory Attention Module (MCMAM) for suppressing background noise and modeling local spatial context. To guide specialized learning, we derive small polyp and edge labels alongside ground truth, enabling MemoryFormer to process them through dedicated branches. The outputs are fused using a Small Polyp Fusion Strategy (SPFS) and an Edge Correction Strategy (ECS) to alleviate over- and under-segmentation. The quantitative results on Kvasir-SEG, CVC-ColonDB, CVC-ClinicDB, CVC-300, and ETIS-Larib yield mean Dice scores of 0.930, 0.818, 0.943, 0.912, and 0.763, respectively, demonstrating strong generalization across diverse polyp segmentation scenarios. Code and datasets are available at: https://github.com/taojlu/PolypSeg.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Image and Vision Computing
工程技术-工程:电子与电气
CiteScore
8.50
自引率
8.50%
发文量
143
审稿时长
7.8 months
期刊介绍:
Image and Vision Computing has as a primary aim the provision of an effective medium of interchange for the results of high quality theoretical and applied research fundamental to all aspects of image interpretation and computer vision. The journal publishes work that proposes new image interpretation and computer vision methodology or addresses the application of such methods to real world scenes. It seeks to strengthen a deeper understanding in the discipline by encouraging the quantitative comparison and performance evaluation of the proposed methodology. The coverage includes: image interpretation, scene modelling, object recognition and tracking, shape analysis, monitoring and surveillance, active vision and robotic systems, SLAM, biologically-inspired computer vision, motion analysis, stereo vision, document image understanding, character and handwritten text recognition, face and gesture recognition, biometrics, vision-based human-computer interaction, human activity and behavior understanding, data fusion from multiple sensor inputs, image databases.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


