加速概率隐私保护医疗记录链接:一种三方MPC方法
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
目的:记录链接对于在现实世界的医疗保健和研究中整合来自多个来源的不同应用的数据至关重要。概率隐私保护记录链接(PPRL)实现了这种集成,同时保护敏感信息免受未经授权的访问,特别是当数据集缺乏精确的标识符时。随着隐私法规的发展和多机构合作在全球范围内的扩展,对有效平衡安全性、准确性和效率的方法的需求不断增长。然而,在大规模记录链接中,确保隐私和可扩展性仍然是一个关键的挑战。方法:提出一种基于安全三方计算(MPC)框架的新型高效PPRL方法。我们的方法允许多方在不暴露其私有输入的情况下计算链接结果,与现有的PPRL解决方案相比,显著提高了链接过程的速度。结果:我们的方法保留了最先进的(SOTA)基于mpc的PPRL方法的连接质量,同时实现了高达14倍的性能提升。例如,在带宽为700 Mbps、延迟为60 ms的实际网络中,将一条记录与包含10,000条记录的数据库相关联只需要8.74 s,而使用SOTA方法需要92.32 s。即使在带宽为100mbps、延迟为60ms的较慢的互联网连接上,连接也需要在28秒内完成,而SOTA方法需要287.96秒。这些结果表明,我们的方法具有显著的可扩展性和效率改进。结论:基于安全三方计算的PPRL方法在保证隐私保护的同时,为大规模记录链接提供了高效、可扩展的解决方案。该方法显示了显著的性能改进,使其成为隐私敏感领域中安全数据集成的有前途的工具。本文章由计算机程序翻译,如有差异,请以英文原文为准。
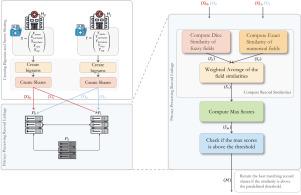
Accelerating probabilistic privacy-preserving medical record linkage: A three-party MPC approach
Objective:
Record linkage is essential for integrating data from multiple sources with diverse applications in real-world healthcare and research. Probabilistic Privacy-Preserving Record Linkage (PPRL) enables this integration occurs, while protecting sensitive information from unauthorized access, especially when datasets lack exact identifiers. As privacy regulations evolve and multi-institutional collaborations expand globally, there is a growing demand for methods that effectively balance security, accuracy, and efficiency. However, ensuring both privacy and scalability in large-scale record linkage remains a key challenge.
Method:
This paper presents a novel and efficient PPRL method based on a secure 3-party computation (MPC) framework. Our approach allows multiple parties to compute linkage results without exposing their private inputs and significantly improves the speed of linkage process compared to existing PPRL solutions.
Result:
Our method preserves the linkage quality of a state-of-the-art (SOTA) MPC-based PPRL method while achieving up to 14 times faster performance. For example, linking a record against a database of 10,000 records takes just 8.74 s in a realistic network with 700 Mbps bandwidth and 60 ms latency, compared to 92.32 s with the SOTA method. Even on a slower internet connection with 100 Mbps bandwidth and 60 ms latency, the linkage completes in 28 s, where as the SOTA method requires 287.96 s. These results demonstrate the significant scalability and efficiency improvements of our approach.
Conclusion:
Our novel PPRL method, based on secure 3-party computation, offers an efficient and scalable solution for large-scale record linkage while ensuring privacy protection. The approach demonstrates significant performance improvements, making it a promising tool for secure data integration in privacy-sensitive sectors.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


