放射性食管炎的多组随机森林毒性模型
IF 3.3
Q2 ONCOLOGY
引用次数: 0
摘要
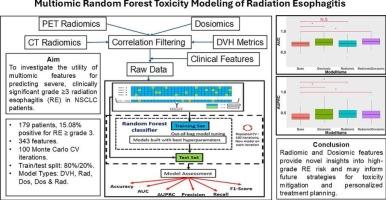
背景和目的放射性食管炎(RE)是非小细胞肺癌(NSCLC)放疗后引起的主要剂量限制性毒性。与传统使用的临床和剂量-体积直方图(DVH)参数相比,多组学特征可能为高级别RE提供额外的预测价值。我们的目的是研究多组学特征在改进稀土毒性预测模型中的应用。材料与方法179例NSCLC患者中,27例(15.08 %)的CTCAE v5.0毒性≥3级RE。共提取了343个基于CT和PET的放射学和剂量学特征。使用以下四组中的临床因素和特征建立了四种毒性预测模型:(a)基础模型(DVH), (b)放射组学,(c)剂量组学,(d)放射组学和剂量组学联合模型。使用随机森林分类器开发模型,该分类器具有100次蒙特卡罗交叉验证迭代和80 %/20 %的训练/测试分割。通过受试者工作特征曲线下面积(AUC)和精确召回率曲线下面积(AUPRC)评价模型的预测性能。ResultsThe AUC和AUPRC值(平均 ± 标准差)4模型类型0.69 ± 0.10/0.42 ±0.12 (基础模型),0.71 ± 0.12/0.48 ±0.13 (radiomic), 0.73 ± 0.10/0.48 ±0.15 (dosiomic)和0.75 ± 0.10/0.49 ± 0.14 (radiomic和dosiomic),分别。采用自举百分位法比较multiomic模型和基本模型的性能指标,结果表明组合模型是性能最好的模型类型。结论所有多组模型均优于基础模型。放射组学-剂量组学联合模型提供了对高级别RE风险的新见解,并可能为未来的毒性缓解策略和个性化治疗计划提供信息。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Multiomic random forest toxicity modeling of radiation esophagitis
Background and purpose
Radiation esophagitis (RE) is a major dose-limiting toxicity resulting from radiotherapy for non-small-cell lung cancer (NSCLC). Multiomic features may provide additional predictive value for high-grade RE compared to traditionally used clinical and dose-volume histogram (DVH) parameters. We aimed to investigate the utility of multiomic features in improving RE toxicity prediction models.
Materials and methods
Of the 179 NSCLC patients considered, 27 patients (15.08 %) were found to have toxicity ≥grade 3 RE per CTCAE v5.0. A total of 343 CT- and PET- based radiomic and dosiomic features were extracted. Four toxicity prediction models were created using clinical factors and features from one of the following groups: (a) base model (DVH), (b) radiomic, (c) dosiomic, and (d) combined radiomic and dosiomic. Models were developed using a random forest classifier with 100 Monte Carlo cross-validation iterations and an 80 %/20 % training/test split. Model predictive performance was evaluated by area under the receiver operating characteristic curve (AUC) and area under the precision-recall curve (AUPRC).
Results
The AUC and AUPRC values (mean ± standard deviation) for the 4 model types were 0.69 ± 0.10/0.42 ± 0.12 (base model), 0.71 ± 0.12/0.48 ± 0.13 (radiomic), 0.73 ± 0.10/0.48 ± 0.15 (dosiomic), and 0.75 ± 0.10/0.49 ± 0.14 (radiomic and dosiomic), respectively. The bootstrap percentile method, which was used to compare performance metrics between multiomic and base models, showed that the combined model was the best performing model type.
Conclusion
All multiomic models outperformed the base model. The combined radiomic-dosiomic model provides novel insights into high-grade RE risk and may inform future strategies for toxicity mitigation and personalized treatment planning.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Physics and Imaging in Radiation Oncology
Physics and Astronomy-Radiation
CiteScore
5.30
自引率
18.90%
发文量
93
审稿时长
6 weeks
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


