在真实的医院环境中使用乳房x光检查预测乳腺癌
IF 6.3
2区 医学
Q1 BIOLOGY
引用次数: 0
摘要
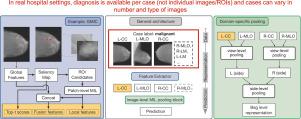
乳房x光检查的乳腺癌预测模型假设对单个图像或感兴趣区域(roi)可用注释,并且每个患者有固定数量的图像。这些假设在真实的医院环境中并不成立,临床医生只提供整个乳房x光检查(病例)的最终诊断。由于真实医院环境中的数据随患者的持续摄入而缩放,而手动注释工作则不需要,因此我们开发了一个病例级乳腺癌预测框架,该框架不需要任何手动注释,并且可以使用医院现成的病例标签进行训练。具体而言,我们提出了一种在斑块和图像级别进行病例级乳腺癌预测的两级多实例学习(MIL)方法,并在两个公共数据集和一个私有数据集上对其进行了评估。我们提出了一种新的域特异性MIL池,观察到乳腺癌可能发生也可能不发生在两侧,而在乳房x光检查中,双侧乳房的图像被作为预防措施。我们提出了一个动态训练过程,用于在每个案例的可变数量的图像上训练我们的MIL框架。我们表明,我们的两级MIL模型可以应用于真实的医院环境,其中只有病例标签,每个病例可用的图像数量可变,与使用图像标签训练的模型相比,性能没有任何损失。仅使用弱(病例级)标签进行训练,它就有能力指出异常位于乳房的哪一侧,乳房x光片视图和视图区域。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Breast cancer prediction using mammography exams for real hospital settings
Breast cancer prediction models for mammography assume that annotations are available for individual images or regions of interest (ROIs), and that there is a fixed number of images per patient. These assumptions do not hold in real hospital settings, where clinicians provide only a final diagnosis for the entire mammography exam (case). Since data in real hospital settings scales with continuous patient intake, while manual annotation efforts do not, we develop a framework for case-level breast cancer prediction that does not require any manual annotation and can be trained with case labels readily available at the hospital. Specifically, we propose a two-level multi-instance learning (MIL) approach at patch and image level for case-level breast cancer prediction and evaluate it on two public and one private dataset. We propose a novel domain-specific MIL pooling observing that breast cancer may or may not occur in both sides, while images of both breasts are taken as a precaution during mammography. We propose a dynamic training procedure for training our MIL framework on a variable number of images per case. We show that our two-level MIL model can be applied in real hospital settings where only case labels, and a variable number of images per case are available, without any loss in performance compared to models trained on image labels. Only trained with weak (case-level) labels, it has the capability to point out in which breast side, mammography view and view region the abnormality lies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


