GraphFusion:使用多尺度图形表示和细胞系上下文对药物协同作用进行综合预测。
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
预测药物组合的协同作用对癌症治疗和药物开发至关重要。准确的预测需要整合多种类型的数据,包括单个药物的分子结构,药物之间可用的协同评分,以及来自不同癌细胞系的基因表达信息。前两种类型包含药物内部或药物之间的多尺度信息,而细胞系则作为药物相互作用的背景。现有的机器学习方法不能充分利用和整合这些信息,导致性能不佳。为了解决这个问题,我们引入了GraphFusion,这是一种将分子图和药物协同作用图与细胞系上下文信息相结合的创新方法。GraphFusion采用能够接受和利用外部信息的新型GCN和graphhormer模块,将这两个层次的图形信息整合在一起。具体来说,分子图将细粒度的结构信息传递给协同图,而协同图将全局药物相互作用数据传递给分子图。此外,细胞系信息被纳入上下文背景。这种全面的集成使GraphFusion能够在O'Neil和NCI-ALMANAC数据集上实现最先进的结果。本文章由计算机程序翻译,如有差异,请以英文原文为准。
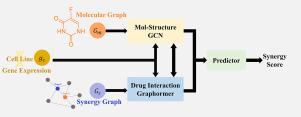
GraphFusion: Integrative prediction of drug synergy using multi-scale graph representations and cell line contexts
Predicting the synergy of drug combinations is crucial for cancer treatment and drug development. Accurate prediction requires the integration of multiple types of data, including molecular structures of individual drugs, available synergy scores between drugs, and gene expression information from different cancer cell lines. The first two types contain multi-scale information within or between drugs, while the cell lines serve as the contextual background for drug interactions. Existing machine learning methods fail to fully utilize and integrate these information, leading to suboptimal performance. To address this issue, we introduce GraphFusion, an innovative approach that combines molecular graphs and drug synergy graphs with cell line contextual information. By employing novel GCN and Graphormer modules capable of accepting and utilizing external information, GraphFusion integrates these two levels of graph information. Specifically, the molecular graphs pass fine-grained structural information to the synergy graphs, while the synergy graphs convey global drug interaction data to the molecular graphs. Additionally, cell line information is incorporated as contextual background. This comprehensive integration enables GraphFusion to achieve state-of-the-art results on the O’Neil and NCI-ALMANAC datasets.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


