超级增强子驱动的ELOVL5通过MYC-SERBP1途径促进T-ALL进展。
IF 3
2区 生物学
Q2 BIOTECHNOLOGY & APPLIED MICROBIOLOGY
引用次数: 0
摘要
急性淋巴细胞白血病(Acute lymphoblastic leukemia, ALL)是儿童最常见的恶性肿瘤,其中t细胞急性淋巴细胞白血病(T-ALL)占10- 25% %。我们确定ELOVL5是一种超级增强子驱动的癌基因,在T-ALL中高度表达,并与较差的总生存率相关。通过对患者样本和细胞模型的H3K27ac ChIP-seq分析,我们证实ELOVL5受超增强子的转录调控。功能研究表明,ELOVL5敲低可抑制T-ALL细胞的增殖并诱导细胞凋亡,无论在体内还是体外。在小鼠异种移植模型中,沉默ELOVL5可减少肿瘤负荷并延长生存期。RNA-seq分析进一步揭示,ELOVL5通过激活MYC信号和上调SERBP1(一种关键的下游效应物)来促进T-ALL的进展。一致地,SERBP1沉默也抑制T-ALL细胞的增殖和诱导凋亡。总的来说,这些发现确定了ELOVL5作为一种超级增强子相关的致癌调节剂,通过ELOVL5- serbp1 - myc轴驱动T-ALL进展,突出了其作为治疗靶点的潜力。本文章由计算机程序翻译,如有差异,请以英文原文为准。
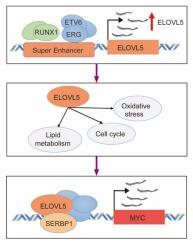
Super-enhancer-driven ELOVL5 promotes T-ALL progression through the MYC-SERBP1 pathway
Acute lymphoblastic leukemia (ALL) is the most common pediatric malignancy, with T-cell acute lymphoblastic leukemia (T-ALL) accounting for 10–25 % of cases. We identified ELOVL5 as a super-enhancer–driven oncogene that is highly expressed in T-ALL and associated with poor overall survival. Through H3K27ac ChIP-seq analysis of patient samples and cellular models, we confirmed that ELOVL5 is transcriptionally regulated by super-enhancers. Functional studies demonstrated that ELOVL5 knockdown suppressed proliferation and induced apoptosis in T-ALL cells, both in vitro and in vivo. In mouse xenograft models, silencing ELOVL5 reduced tumor burden and prolonged survival. RNA-seq analysis further revealed that ELOVL5 promotes T-ALL progression by activating MYC signaling and upregulating SERBP1, a critical downstream effector. Consistently, SERBP1 silencing also inhibited proliferation and induced apoptosis in T-ALL cells. Collectively, these findings establish ELOVL5 as a super-enhancer–associated oncogenic regulator that drives T-ALL progression through the ELOVL5–SERBP1–MYC axis, highlighting its potential as a therapeutic target.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Genomics
生物-生物工程与应用微生物
CiteScore
9.60
自引率
2.30%
发文量
260
审稿时长
60 days
期刊介绍:
Genomics is a forum for describing the development of genome-scale technologies and their application to all areas of biological investigation.
As a journal that has evolved with the field that carries its name, Genomics focuses on the development and application of cutting-edge methods, addressing fundamental questions with potential interest to a wide audience. Our aim is to publish the highest quality research and to provide authors with rapid, fair and accurate review and publication of manuscripts falling within our scope.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


