基于路径引导和自适应负采样增强的药物重新定位。
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
目的:药物重新定位在加快药物发现过程中起着关键作用。计算方法的快速发展为预测药物-疾病关联(DDAs)开辟了新的途径。尽管现有方法取得了进步,但诸如对异质生物网络中多种关系的探索不足和阴性样本质量不足等挑战仍然存在。方法:在本研究中,我们引入了一种新的药物重新定位框架DRMGNE,该框架利用元路径引导学习和自适应负增强来预测DDA。DRMGNE首先使用自编码器提取基于相似矩阵的语义特征。随后,设计了一套全面的元路径来生成子图,并利用图卷积网络提取反映拓扑结构的丰富节点表示。此外,采用自适应负增强策略提高负样本的质量,保证均衡学习。结果:实验评估表明,DRMGNE在三个基准数据集上优于最先进的算法。此外,案例研究和分子对接验证进一步强调了其在促进药物发现和加速药物再利用方面的潜力。结论:DRMGNE是一种新的DDA预测框架,它利用了基于元路径的引导和自适应负增强。在基准数据集上的实验显示了优于现有方法的性能,强调了其在药物发现中的潜在影响。本文章由计算机程序翻译,如有差异,请以英文原文为准。
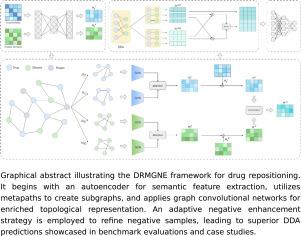
Drug repositioning with metapath guidance and adaptive negative sampling enhancement
Objective:
Drug repositioning plays a pivotal role in expediting the drug discovery pipeline. The rapid development of computational methods has opened new avenues for predicting drug-disease associations (DDAs). Despite advancements in existing methodologies, challenges such as insufficient exploration of diverse relationships in heterogeneous biological networks and inadequate quality of negative samples have persisted.
Methods:
In this study, we introduce DRMGNE, a novel drug repositioning framework that harnesses metapath-guided learning and adaptive negative enhancement for DDA prediction. DRMGNE initiates with an autoencoder to extract semantic features based on similarity matrices. Subsequently, a comprehensive set of metapaths is designed to generate subgraphs, and graph convolutional networks are utilized to extract enriched node representations reflecting topological structures. Furthermore, the adaptive negative enhancement strategy is employed to improve the quality of negative samples, ensuring balanced learning.
Results:
Experimental evaluations demonstrate that DRMGNE outperforms state-of-the-art algorithms across three benchmark datasets. Additionally, case studies and molecular docking validations further underscore its potential in facilitating drug discovery and accelerating drug repurposing efforts.
Conclusion:
DRMGNE is a novel framework for DDA prediction that leverages metapath-based guidance and adaptive negative enhancement. Experiments on benchmark datasets show superior performance over existing methods, underscoring its potential impact in drug discovery.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


