埃博拉病毒转录调控信号在两种结构构象之间平衡以影响VP30的结合。
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
埃博拉病毒(EBOV)基因组的转录是病毒生命周期中的一个主要检查点。改变转录调控信号(TRS),即在EBOV 3'先导序列中形成的RNA发夹,以依赖vp30的方式影响转录。目前尚不清楚这种机制是如何发生的。在这里,我们发现TRS主要在两个不同的结构之间保持平衡。突变研究表明,整个发夹结构在指导VP30结合和调节EBOV转录活性方面都很重要,而这个环是主要的结构元件。在此过程中,我们强调TRS结构是EBOV转录的关键决定因素。本文章由计算机程序翻译,如有差异,请以英文原文为准。
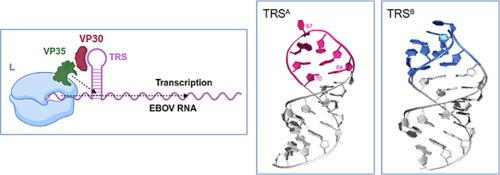
The Ebolavirus Transcription Regulatory Signal Equilibrates Between Two Structural Conformations to Affect VP30 Binding
The transcription of ebolavirus (EBOV) genomes is a major checkpoint in the viral life cycle. Altering the transcription regulatory signal (TRS), an RNA hairpin that forms in the 3′ leader sequence of EBOV, affects transcription in a VP30-dependent manner. How this mechanistically occurs is currently unknown. Here, we find that the TRS primarily equilibrates between two structures that differ in their loops. Mutational studies demonstrate that the entire hairpin structure is important in directing both VP30 binding and regulating EBOV transcriptional activity, with the loop being the principal structural element. In doing so, we highlight that the TRS structure is a critical determinant of EBOV transcription.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


