Silva模式引导的蛋白质组学揭示了子宫颈腺癌中协调的肿瘤-基质重塑
IF 4.1
2区 医学
Q1 OBSTETRICS & GYNECOLOGY
引用次数: 0
摘要
目的探讨宫颈内膜腺癌(ECA)微环境中蛋白质组的变化。方法32例ECA分别为Silva A (n10)、Silva B (n11)和Silva C (n11)肿瘤。肿瘤进行激光显微解剖,富集肿瘤和邻近的基质细胞,然后进行多重定量蛋白质组学分析。比较分析采用分层聚类和差分统计与LIMMA进行,包括通路改变的预测和蛋白质定位的优先级作为假定的药物靶点。结果定量蛋白质组学分析在所有微室样品中鉴定出7300个蛋白质。无监督分析显示肿瘤和基质种群之间存在明显差异,并将Silva C基质与Silva模式A/B区分开来。肿瘤分裂成簇,富集线粒体代谢、免疫信号或细胞外基质通路;基质形成互补簇,以基质重塑、免疫或细胞周期程序为特征。与模式A和B相比,Silva C肿瘤和基质细胞表现出大量的蛋白质改变,包括肿瘤-基质亚群之间共有的候选蛋白(CPA3, NNMT),突出了微环境的互易性。很强的肿瘤-基质相关性(Spearman ρ > 0.83)强调了协调重塑,并且在Silva C病例中基质蛋白的改变丰富,表明在这些肿瘤中有明显的免疫激活。Silva A和C肿瘤之间改变的候选药物靶点包括PDGFRB、CDK4、EGFR、MAP2K1/2和CEACAM5。结论基于观察到的蛋白质组改变,silva C ECA表现出独特的免疫和基质中心肿瘤基质轴。肿瘤微环境分解蛋白质组学鉴定了Silva模式特异性生物标志物和治疗脆弱性,保证了功能和临床验证。本文章由计算机程序翻译,如有差异,请以英文原文为准。
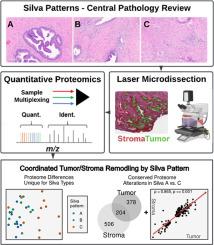
Silva pattern-guided proteomics reveals coordinated tumor-stroma remodeling in cervical adenocarcinoma
Objective
Characterize proteome alterations within the tumor microenvironment of endocervical adenocarcinoma (ECA) stratified by Silva patterns.
Methods
Pathology review assigned 32 ECA as Silva A (n10), Silva B (n11) and Silva C (n11) tumors. Tumors underwent laser-microdissection enrichment of tumor and adjacent stromal cells followed by multiplexed, quantitative proteomic analyses. Comparative analyses were performed using hierarchical clustering and differential statistics with LIMMA and included prediction of pathway alterations and prioritization of proteins mapping as putative drug targets.
Results
Quantitative proteomic analyses identified 7300 proteins across all microcompartment samples. Unsupervised analyses showed stark differences between tumor and stroma populations and distinguished Silva C stroma from Silva patterns A/B. Tumors partitioned into clusters enriched for mitochondrial metabolism, immune signaling, or extracellular-matrix pathways; stroma formed complementary clusters characterized by matrix remodeling, immunity, or cell-cycle programs. Compared to patterns A and B, Silva C tumor and stroma cells exhibited large numbers of protein alterations and included candidates shared between tumor–stroma subpopulations (CPA3, NNMT), highlighting microenvironment reciprocity. Strong tumor-stroma correlations (Spearman ρ > 0.83) underscored coordinated remodeling, and stromal protein alterations enriched in Silva C cases, suggested pronounced immune activation in these tumors. Candidate drug targets altered between Silva A and C tumors included PDGFRB, CDK4, EGFR, MAP2K1/2, and CEACAM5.
Conclusions
Silva C ECA exhibits a distinctive immune- and matrix-centric tumor–stroma axis based on proteome alterations observed. Tumor microenvironment-resolved proteomics identified Silva pattern-specific biomarkers and therapeutic vulnerabilities that warrant functional and clinical validation.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Gynecologic oncology
医学-妇产科学
CiteScore
8.60
自引率
6.40%
发文量
1062
审稿时长
37 days
期刊介绍:
Gynecologic Oncology, an international journal, is devoted to the publication of clinical and investigative articles that concern tumors of the female reproductive tract. Investigations relating to the etiology, diagnosis, and treatment of female cancers, as well as research from any of the disciplines related to this field of interest, are published.
Research Areas Include:
• Cell and molecular biology
• Chemotherapy
• Cytology
• Endocrinology
• Epidemiology
• Genetics
• Gynecologic surgery
• Immunology
• Pathology
• Radiotherapy
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


