联邦学习满足少次学习:一种基于投票集成的跨非iid数据分布的花椰菜叶病分类组合方法
IF 4.5
Q2 COMPUTER SCIENCE, THEORY & METHODS
引用次数: 0
摘要
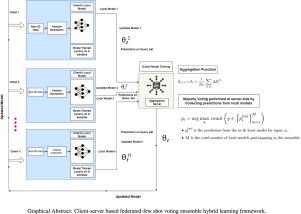
近年来,花椰菜(Brassica oleracea var. botrytis)叶片病害的分类在农业研究中备受关注,特别是在优化产量和加强病害管理方面。传统的机器学习方法通常依赖于大型、分布良好的数据集,由于不同地区和农场的数据特征不同,这些数据集很难获得。为了解决这一挑战,我们提出了一个新的框架,该框架将联邦学习(FL)与Few-Shot学习(FSL)集成在一起,即使在数据分布不相同的环境中也能进行稳健的花椰菜叶片病害检测。我们的混合方法在任务级别进行泛化,而不需要大量未标记的数据集,这与传统的FL方法不同,后者严重依赖数据增强或半监督技术来弥补数据不足。few - shot Learning使每个客户只需少量样本就能更容易地适应新的疾病类别。这使得混合框架在数据不是独立和相同分布(IID)的情况下更加灵活和高效。在我们的研究中,我们使用了五个客户端,每个客户端在n-way k-shot配置中都有自己的支持和查询集,其中每个客户端在一小组标记数据上进行训练,并在未见过的数据上评估模型。联邦学习促进了这些客户端之间的协作和分散训练,而通过爬行动物元学习算法实现的Few-Shot学习允许每个客户端在有限的样本下有效地适应新的类。此外,我们通过集成投票提高了预测精度,其中组合了来自多个预训练深度学习模型(VGG16, ResNet50V2, Xception, DenseNet169和MobileNetV2)的预测。集成投票将单个模型的预测聚合在一起,并选择所有模型中最常见的类预测,从而提高整体分类性能。该集成机制在每个联邦轮之后实现,其中模型权重根据本地客户训练进行聚合和更新,然后进行疾病分类的最终集成决策。此外,我们对我们的框架进行了消融研究,以评估每个组件(FL, FSL和投票集合)的贡献。实验结果表明,该集成模型在2弹、3弹和4弹配置下的测试精度分别为95%、97%和100%,验证了该框架的有效性。结果表明,我们的Fed-FSL混合框架与集成投票相结合,即使在异构环境中也能提供准确的疾病分类,为精准农业和智能农业系统提供了可扩展和适应性强的解决方案。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Federated learning meets few-shot learning: A voting ensemble based combined approach to cauliflower leaf disease classification across Non-IID data distributions
In recent years, the classification of cauliflower (Brassica oleracea var. botrytis) leaf diseases has gained significant attention within agricultural research, particularly for optimizing yield and enhancing disease management. Traditional machine learning methods often rely on large, well-distributed datasets, which are difficult to obtain due to diverse data characteristics across regions and farms. To address this challenge, we proposed a novel framework that integrates Federated Learning (FL) with Few-Shot Learning (FSL) for robust cauliflower leaf disease detection, even in environments with non-identical data distributions. Our hybrid method generalizes at the task level without needing large unlabeled datasets, unlike traditional FL methods that rely heavily on data augmentation or semi-supervised techniques to make up for a lack of data. Few-Shot Learning makes it easier for each client to adapt to new disease classes with only a few samples. This makes the hybrid framework more flexible and efficient in situations where the data is not independent and identically distributed (IID). In our study, we utilized five clients, each having their own support and query sets in an n-way k-shot configuration, where each client trains on a small set of labeled data and evaluates the model on unseen data. Federated Learning facilitates collaborative, decentralized training among these clients, while Few-Shot Learning, implemented through the Reptile meta-learning algorithm, allows each client to efficiently adapt to new classes with limited samples. Moreover, we enhance prediction accuracy through ensemble voting, where predictions from multiple pre-trained deep learning models (VGG16, ResNet50V2, Xception, DenseNet169, and MobileNetV2) are combined. The ensemble voting aggregates the individual model predictions, and the most common class prediction across all models is selected, improving the overall classification performance. This ensemble mechanism is implemented after each federated round, where the model weights are aggregated and updated based on local client training, followed by a final ensemble decision for disease classification. Additionally, we have performed an ablation study on our framework to evaluate the contribution of each component (FL, FSL, and voting ensemble). Experimental results show that the ensemble model achieves an test accuracy of 95% for 2-shot, 97% for 3-shot and 4-shot, and 100% for 5-shot configurations, demonstrating the effectiveness of the framework. The results demonstrate that our Fed-FSL hybrid framework, combined with ensemble voting, provides accurate disease classification even in heterogeneous environments, offering a scalable and adaptable solution for precision agriculture and smart farming systems.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


