基于方向约束扩散的单细胞微扰响应预测Schrödinger桥
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
目的预测单细胞水平对外部扰动的转录反应对于理解基因调控网络、药物发现和个性化干预至关重要。扰动条件的指数增长造成数据稀疏性,使得难以捕获动态响应并需要计算建模。方法我们提出了方向约束扩散Schrödinger桥(DC-DSB),这是一个生成框架,通过最小化路径空间KL散度来学习无扰动和后扰动分布之间的概率轨迹。为了增强条件控制,DC-DSB集成了由实验变量和生物先验知识派生的层次表示。我们进一步引入了一种方向约束的条件反射策略,该策略沿着生物相关的扰动轨迹注入条件信号,从而提高建模质量和训练稳定性。结果dc - dsb在基线上提高了表达预测的准确性和对未见组合的泛化。通过模拟扰动下的动态表达轨迹和共表达结构,DC-DSB能够发现协同和拮抗基因相互作用,并支持调控途径的逐步重建。结论dc - dsb为单细胞微扰建模提供了生物学一致性和可推广的框架。其基于轨迹和条件感知的结构克服了静态映射的局限性,促进了基因调控和药物发现的下游分析。本文章由计算机程序翻译,如有差异,请以英文原文为准。
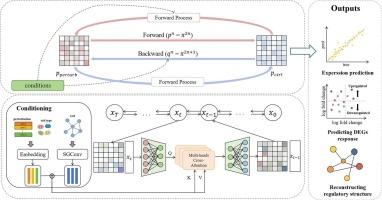
Prediction of Single-Cell perturbation response based on Direction-Constrained diffusion Schrödinger Bridge
Objective
Predicting transcriptional responses to external perturbations at the single-cell level is essential for understanding gene regulatory networks, drug discovery, and personalized interventions. The exponential increase in perturbation conditions creates data sparsity, making it difficult to capture dynamic responses and necessitating computational modeling.
Methods
We present Direction-Constrained Diffusion Schrödinger Bridge (DC-DSB), a generative framework that learns probabilistic trajectories between unperturbed and post-perturbation distributions by minimizing path-space KL divergence. To enhance conditional control, DC-DSB integrates hierarchical representations derived from experimental variables and biological prior knowledge. We further introduce a direction-constrained conditioning strategy that injects condition signals along the biologically relevant perturbation trajectory, thereby improving modeling quality and training stability.
Results
DC-DSB improves expression prediction accuracy and generalization to unseen combinations over baselines. By modeling dynamic expression trajectories and co-expression structures under perturbation, DC-DSB enables the discovery of synergistic and antagonistic gene interactions and supports the progressive reconstruction of regulatory pathways.
Conclusion
DC-DSB provides a biologically consistent and generalizable framework for single-cell perturbation modeling. Its trajectory-based and condition-aware architecture overcomes the limitations of static mappings and facilitates downstream analyses in gene regulation and drug discovery.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


