揭示癌症干细胞标记网络:超图方法
IF 3.1
4区 生物学
Q2 BIOLOGY
引用次数: 0
摘要
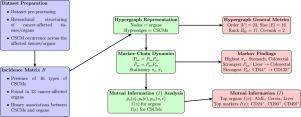
我们提出了一个新的计算框架利用超图理论来分析跨多个器官的癌症干细胞标记物(CSCMs)。超图提供了CSCM共表达模式的鲁棒表示,比传统的基于图的方法更全面地捕获了它们复杂的多器官关系。通过整合互信息分析和马尔可夫模型,我们确定了驱动肿瘤异质性和转移的关键标志物,并提供了对其相互依赖性的详细见解。这种方法建立了超图作为模拟癌症进展和转移动力学的强大计算工具,有助于理解复杂的生物系统并支持靶向治疗策略的发展。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Unveiling cancer stem cell marker networks: A hypergraph approach
We propose a novel computational framework leveraging hypergraph theory to analyse cancer stem cell markers (CSCMs) across multiple organs. Hypergraphs provide a robust representation of CSCM co-expression patterns, capturing their complex multi-organ relationships more comprehensively than traditional graph-based methods. By integrating mutual information analysis and Markov models, we identify key markers driving tumour heterogeneity and metastasis, offering detailed insights into their interdependencies. This approach establishes hypergraphs as a computationally powerful tool to model cancer progression and metastatic dynamics, contributing to the understanding of complex biological systems and supporting the development of targeted therapeutic strategies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computational Biology and Chemistry
生物-计算机:跨学科应用
CiteScore
6.10
自引率
3.20%
发文量
142
审稿时长
24 days
期刊介绍:
Computational Biology and Chemistry publishes original research papers and review articles in all areas of computational life sciences. High quality research contributions with a major computational component in the areas of nucleic acid and protein sequence research, molecular evolution, molecular genetics (functional genomics and proteomics), theory and practice of either biology-specific or chemical-biology-specific modeling, and structural biology of nucleic acids and proteins are particularly welcome. Exceptionally high quality research work in bioinformatics, systems biology, ecology, computational pharmacology, metabolism, biomedical engineering, epidemiology, and statistical genetics will also be considered.
Given their inherent uncertainty, protein modeling and molecular docking studies should be thoroughly validated. In the absence of experimental results for validation, the use of molecular dynamics simulations along with detailed free energy calculations, for example, should be used as complementary techniques to support the major conclusions. Submissions of premature modeling exercises without additional biological insights will not be considered.
Review articles will generally be commissioned by the editors and should not be submitted to the journal without explicit invitation. However prospective authors are welcome to send a brief (one to three pages) synopsis, which will be evaluated by the editors.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


