天然质谱法筛选RNA适体的小分子研究
IF 4.7
2区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
用小分子靶向RNA被认为是一种可行的新疗法,具有极大扩大可药物化学空间的潜力。然而,有意靶向RNA的实际考虑在很大程度上仍在发展中,包括识别与RNA结合的小分子的最佳方法。原生质谱(nMS)是一种有价值的生物物理筛选方法,用于鉴定蛋白质靶点的小分子命中,但用于寡核苷酸的程度要小得多。本文应用nMS分析了两种氨基糖苷RNA适配体与其同源配体、相关氨基糖苷配体和FDA批准药物的多种小分子文库的结合。nMS证实了同源配体的结合,并允许半定量地确定所有被测化合物的结合强度和选择性。nMS库筛选还发现了新的粘结剂。这项工作表明,nMS可以应用于RNA适体的生物物理筛选,并且有潜力在更广泛的RNA靶向药物发现中发展为正交筛选技术,但是与RNA的物理化学性质所带来的具体挑战相关的局限性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
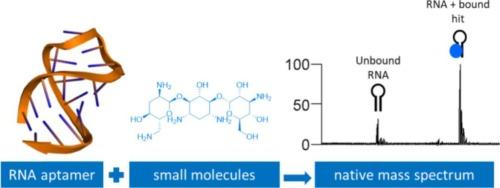
Small molecule screening of RNA aptamers using native mass spectrometry
The intentional targeting of RNA with small molecules is recognized as a viable pathway to new therapeutics with potential to vastly expand the druggable chemical space. The practical considerations to deliberately target RNA are however still largely under development, including optimal methods to identify small molecules for binding to RNA. Native mass spectrometry (nMS) is established as a valuable biophysical screening method for identifying small molecule hits for protein targets but has been used to a much lesser extent with oligonucleotides. Herein we applied nMS to the analysis of binding of two aminoglycoside RNA aptamers with their cognate ligands, related aminoglycoside ligands and a diverse small molecule library of FDA approved drugs. nMS confirmed cognate ligand binding and allowed semi-quantitation of binding strength and selectivity to be determined across all compounds tested. nMS library screening also identified novel binders. This work demonstrates that nMS can be applied for biophysical screening of RNA aptamers and has potential to be developed as an orthogonal screening technology within broader RNA-targeting drug discovery, however with limitations related to the specific challenges presented by the physicochemical properties of RNA.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Bioorganic Chemistry
生物-生化与分子生物学
CiteScore
9.70
自引率
3.90%
发文量
679
审稿时长
31 days
期刊介绍:
Bioorganic Chemistry publishes research that addresses biological questions at the molecular level, using organic chemistry and principles of physical organic chemistry. The scope of the journal covers a range of topics at the organic chemistry-biology interface, including: enzyme catalysis, biotransformation and enzyme inhibition; nucleic acids chemistry; medicinal chemistry; natural product chemistry, natural product synthesis and natural product biosynthesis; antimicrobial agents; lipid and peptide chemistry; biophysical chemistry; biological probes; bio-orthogonal chemistry and biomimetic chemistry.
For manuscripts dealing with synthetic bioactive compounds, the Journal requires that the molecular target of the compounds described must be known, and must be demonstrated experimentally in the manuscript. For studies involving natural products, if the molecular target is unknown, some data beyond simple cell-based toxicity studies to provide insight into the mechanism of action is required. Studies supported by molecular docking are welcome, but must be supported by experimental data. The Journal does not consider manuscripts that are purely theoretical or computational in nature.
The Journal publishes regular articles, short communications and reviews. Reviews are normally invited by Editors or Editorial Board members. Authors of unsolicited reviews should first contact an Editor or Editorial Board member to determine whether the proposed article is within the scope of the Journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


