一些新设计的配体在Epstein Barr EBNA1/DNA结合域中的行为的MD模拟和结合自由能计算
IF 2.9
4区 工程技术
Q1 MULTIDISCIPLINARY SCIENCES
引用次数: 0
摘要
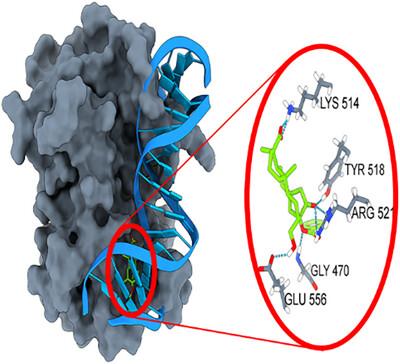
爱泼斯坦-巴尔核抗原1需要DNA结合才能发挥作用。由于目前还没有美国食品和药物管理局批准的对抗eb - barr病毒的药物,本研究旨在研究以前设计的配体在动态环境中的结合特性。因此,除了五种配体外,研究还包括载脂蛋白和dna -蛋白质复合物的分子动力学研究。配体-蛋白质复合物的RMSD图显示,当系统达到稳定状态时,配体-蛋白质复合物中蛋白质的延伸环比dna -蛋白质复合物更灵活。其中,B288的结合自由能值为- 58.63 kcal mol - 1,结合自由能值最高,B183次之,为- 58.24 kcal mol - 1。定义设计的配体B175、B183和B288比从ZINC15数据库中获得的配体Z067和Z108更有利于阻断dna -蛋白结合。研究发现,通过靶向研究的蛋白质结合位点,可以阻止DNA与EBNA1抗原的结合。此外,分子动力学研究表明,紧密包裹DNA的蛋白质延伸环被配体扭曲,这也可能阻碍DNA与蛋白质的结合。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Investigation of the Behaviors of Some Newly Designed Ligands in the Epstein Barr EBNA1/DNA Binding Domain by MD Simulations and Binding Free Energy Calculations
The Epstein-Barr nuclear antigen 1 requires DNA binding to function. Since no Food and Drug Administration-approved drugs exist to combat with Ebstein-Barr virus, the study aims to investigate the binding properties of formerly designed ligands in dynamic environment. Thus, in addition to five ligands, study includes molecular dynamics studies of apo-protein and DNA-protein complex. The RMSD plots of ligand-protein complexes revealed that while systems are reached stable states the extended loops of protein are more flexible in ligand-protein complexes than in DNA-protein complex. Among the ligands, B288 showed best binding properties with a binding free energy value of −58.63 kcal mol−1 which is followed by B183 with the value of −58.24 kcal mol−1. It is defined that designed ligands B175, B183, and B288 are more favorable in blocking DNA-protein binding than Z067 and Z108 ligands obtained from ZINC15 database. The study revealed that the binding of DNA to the EBNA1 antigen can be prevented by targeting the studied binding site of protein. Moreover, the molecular dynamics studies demonstrated the extended loop of protein which wraps up DNA closely is distorted by the ligands, which may also hamper binding of DNA to protein.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Advanced Theory and Simulations
Multidisciplinary-Multidisciplinary
CiteScore
5.50
自引率
3.00%
发文量
221
期刊介绍:
Advanced Theory and Simulations is an interdisciplinary, international, English-language journal that publishes high-quality scientific results focusing on the development and application of theoretical methods, modeling and simulation approaches in all natural science and medicine areas, including:
materials, chemistry, condensed matter physics
engineering, energy
life science, biology, medicine
atmospheric/environmental science, climate science
planetary science, astronomy, cosmology
method development, numerical methods, statistics
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


