使用异构的真实生存数据开发联邦时间到事件评分。
IF 6.3
2区 医学
Q1 BIOLOGY
引用次数: 0
摘要
目的:生存分析在许多医疗保健应用中是一个基本组成部分,其中确定患者特定事件(如某种疾病的发作或死亡)的时间对临床决策至关重要。评分系统被广泛用于快速有效的风险预测。然而,现有的构建生存分数的方法假设数据来源于单一来源,这在与多个数据所有者合作时提出了隐私挑战。材料和方法:我们提出了一种新的框架,用于为多站点生存结果构建联邦评分系统,确保隐私和通信效率。我们将我们的方法应用于来自新加坡和美国急诊科的异质生存数据站点。此外,我们在每个站点独立开发了本地分数。结果:在每个参与者站点的测试数据集中,我们提出的联邦评分系统始终优于所有本地模型,在接受者工作特征曲线(iAUC)值下具有更高的集成面积,最大改进为11.6%。此外,联邦分数与时间相关的AUC(t)值显示出优于本地分数的优势,在大多数时间点上显示出更窄的置信区间(ci)。讨论:通过我们提出的方法开发的模型显示出良好的局部性能,并有望用于未来的医疗保健研究。参与我们提出的联邦评分模型训练的站点可以开发具有增强预测准确性和效率的生存模型。结论:本研究证明了我们的隐私保护联合生存评分生成框架的有效性及其对现实世界异构生存数据的适用性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
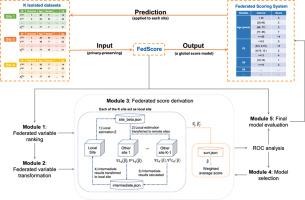
Developing federated time-to-event scores using heterogeneous real-world survival data
Objective
Survival analysis serves as a fundamental component in numerous healthcare applications, where the determination of the time to specific events (such as the onset of a certain disease or death) for patients is crucial for clinical decision-making. Scoring systems are widely used for swift and efficient risk prediction. However, existing methods for constructing survival scores presume that data originates from a single source, posing privacy challenges in collaborations with multiple data owners.
Materials and methods
We propose a novel framework for building federated scoring systems for multi-site survival outcomes, ensuring both privacy and communication efficiency. We applied our approach to sites with heterogeneous survival data originating from emergency departments in Singapore and the United States. Additionally, we independently developed local scores at each site.
Results
In testing datasets from each participant site, our proposed federated scoring system consistently outperformed all local models, evidenced by higher integrated area under the receiver operating characteristic curve (iAUC) values, with a maximum improvement of 11.6 %. Additionally, the federated score's time-dependent AUC(t) values showed advantages over local scores, exhibiting narrower confidence intervals (CIs) across most time points.
Discussion
The model developed through our proposed method showed good local performance and is promising for future healthcare research. Sites participating in our proposed federated scoring model training can develop survival models with enhanced prediction accuracy and efficiency.
Conclusion
This study demonstrates the effectiveness of our privacy-preserving federated survival score generation framework and its applicability to real-world heterogeneous survival data.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computers in biology and medicine
工程技术-工程:生物医学
CiteScore
11.70
自引率
10.40%
发文量
1086
审稿时长
74 days
期刊介绍:
Computers in Biology and Medicine is an international forum for sharing groundbreaking advancements in the use of computers in bioscience and medicine. This journal serves as a medium for communicating essential research, instruction, ideas, and information regarding the rapidly evolving field of computer applications in these domains. By encouraging the exchange of knowledge, we aim to facilitate progress and innovation in the utilization of computers in biology and medicine.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


