UHRF1通过催化HMGA2启动子甲基化抑制巨噬细胞M1极化,从而改善肝硬化。
IF 5.6
2区 医学
Q1 PHARMACOLOGY & PHARMACY
引用次数: 0
摘要
目前,抑制巨噬细胞M1极化和HSC活化被认为是肝硬化的潜在治疗策略。泛素样PHD和环指结构域1 (UHRF1)是一种重要的表观遗传调节剂,参与维持DNA甲基化和染色质结构。据报道,高迁移率组A蛋白2 (HMGA2)的敲低可改善非酒精性脂肪肝的肝脏炎症和纤维化。在这里,我们研究了uhrf1介导的HMGA2启动子甲基化在肝硬化进程中的复杂作用。对肝硬化患者肝组织进行转录组测序,并通过每周2次、连续4周腹腔注射四氯化碳(CCl4)建立肝硬化小鼠模型。用过表达的UHRF1腺病毒转染脂多糖(LPS)诱导的RAW264.7,并以细胞上清作为培养肝星状细胞(hsc)的条件培养基。值得注意的是,在体内实验中,UHRF1表达的增强减轻了小鼠肝脏的纤维化和炎症。在体外实验中,上调UHRF1可抑制ccl4刺激的巨噬细胞M1极化和HSC活化。此外,MSP分析显示,UHRF1过表达后,HMGA2启动子-599/2-450位点的甲基化增强。更有趣的是,ChIP和Co-IP结果澄清了UHRF1与DNA甲基转移酶1 (DNMT1)相互作用,从而结合到HMGA2启动子并促进其甲基化。HMGA2水平升高可消除UHRF1上调对巨噬细胞促炎基因和造血干细胞胶原编码基因mRNA表达的抑制作用。总的来说,UHRF1通过催化HMGA2启动子甲基化来减弱HMGA2的表达,从而阻断巨噬细胞M1极化,减轻肝硬化。本文章由计算机程序翻译,如有差异,请以英文原文为准。
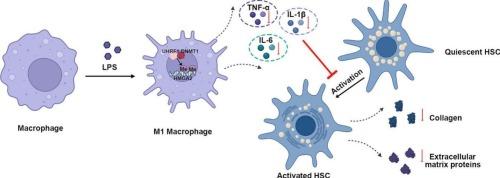
UHRF1 ameliorates liver cirrhosis through suppressing macrophage M1 polarization by catalyzing HMGA2 promoter methylation
Currently, suppressing macrophage M1 polarization and HSC activation are recognized as potential therapeutic strategies for liver cirrhosis. Ubiquitin-like with PHD and RING Finger Domains 1 (UHRF1) is a crucial epigenetic modulator implicated in maintaining DNA methylation and chromatin structure. Knockdown of high mobility group A protein 2 (HMGA2) was reported to ameliorate hepatic inflammation and fibrosis in nonalcoholic fatty liver disease. Here, the elaborate effect of UHRF1-mediated HMGA2 promoter methylation was investigated during the progression of liver cirrhosis. Transcriptomic sequencing of liver tissues of cirrhotic patients was carried out and a mouse model with liver cirrhosis was established by intraperitoneal injection of carbon tetrachloride (CCl4) twice weekly for four consecutive weeks. Lipopolysaccharide (LPS)-induced RAW264.7 were transduced with overexpressed UHRF1 adenovirus and cell supernatant was applied as the conditional medium to cultivate hepatic stellate cells (HSCs). Notably, in vivo experiments, enhanced UHRF1 expression alleviated fibrosis and inflammation of mouse livers. In vitro experiments, upregulated UHRF1 suppressed CCl4-stimulated macrophage M1 polarization and HSC activation. Moreover, MSP assay revealed that methylation was enhanced on HMGA2 promoter at −599/2−450 site after UHRF1 overexpression. More intriguingly, ChIP and Co-IP results clarified that UHRF1 interacted with DNA methyltransferase 1 (DNMT1), thereby bound to HMGA2 promoter and facilitated its methylation. Furthermore, increased HMGA2 level abolished the inhibitory effects of UHRF1 upregulation on the mRNA expression of pro-inflammatory genes of macrophages and genes encoding collagen of HSCs. Collectively, UHRF1 weakened HMGA2 expression by catalyzing its promoter methylation, thereby blocked macrophage M1 polarization and mitigated liver cirrhosis.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Biochemical pharmacology
医学-药学
CiteScore
10.30
自引率
1.70%
发文量
420
审稿时长
17 days
期刊介绍:
Biochemical Pharmacology publishes original research findings, Commentaries and review articles related to the elucidation of cellular and tissue function(s) at the biochemical and molecular levels, the modification of cellular phenotype(s) by genetic, transcriptional/translational or drug/compound-induced modifications, as well as the pharmacodynamics and pharmacokinetics of xenobiotics and drugs, the latter including both small molecules and biologics.
The journal''s target audience includes scientists engaged in the identification and study of the mechanisms of action of xenobiotics, biologics and drugs and in the drug discovery and development process.
All areas of cellular biology and cellular, tissue/organ and whole animal pharmacology fall within the scope of the journal. Drug classes covered include anti-infectives, anti-inflammatory agents, chemotherapeutics, cardiovascular, endocrinological, immunological, metabolic, neurological and psychiatric drugs, as well as research on drug metabolism and kinetics. While medicinal chemistry is a topic of complimentary interest, manuscripts in this area must contain sufficient biological data to characterize pharmacologically the compounds reported. Submissions describing work focused predominately on chemical synthesis and molecular modeling will not be considered for review.
While particular emphasis is placed on reporting the results of molecular and biochemical studies, research involving the use of tissue and animal models of human pathophysiology and toxicology is of interest to the extent that it helps define drug mechanisms of action, safety and efficacy.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


