泰国牛和亚洲水牛的嗜血支原体:揭示全球分布的序列类型和区域遗传多样性
IF 1.7
Q3 PARASITOLOGY
Current research in parasitology & vector-borne diseases
Pub Date : 2025-01-01
DOI:10.1016/j.crpvbd.2025.100318
引用次数: 0
摘要
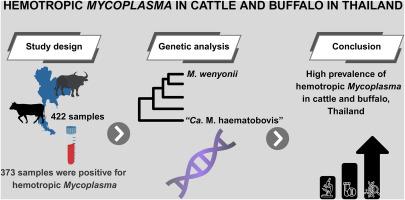
嗜血支原体,特别是温氏支原体和“血牛候选支原体”,越来越被认为是牛和水牛中具有重要兽医意义的新媒介传播病原体。由于其复杂的流行病学和对其致病机制的了解有限,大型反刍动物的血浆感染经常被诊断和报告不足,特别是在资源有限的环境中。本研究调查了2016年至2023年在泰国7个省份采集的牛和水牛血液样本中嗜血支原体物种的遗传多样性和系统发育关系。共分析了422份样本(214份来自牛,208份来自水牛),其中373份嗜血性支原体DNA检测呈阳性。对16S和23S rRNA基因序列进行系统发育分析,发现两个不同的分支分别对应于M. wenyonii和Ca. M. haematobovis。大多数序列与来自多个国家的温氏支原体菌株聚集在一起,而较小的子集与“血牛支原体”菌株聚集在一起。wenyonii M. 16S rRNA基因序列的种内相似性为97.29% ~ 100%,而“Ca. M. haematobovis”序列的种内相似性为99.75% ~ 100%。种间相似度为83.48% ~ 84.34%。wenyonii和Ca. M. haematobovis的23S rRNA基因序列的种内相似度分别为90.94% ~ 100%和99.53%;该基因的种间相似性为76.69% ~ 77.34%。我们鉴定出文氏分枝杆菌的11个核苷酸序列类型(ntSTs),其中ntST10分布最广泛,在古巴、泰国、日本和巴西的分离株中共有。本研究中检测到的单一“Ca. M. haematobovis”序列类型与来自古巴、马来西亚、巴西、土耳其、吉尔吉斯斯坦和日本的序列聚集在ntST7中。这些发现突出了泰国嗜血血支原体物种丰富的遗传多样性和广泛的地理分布,并强调了分子筛选对管理牲畜种群中病原体传播风险的至关重要性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Hemotropic Mycoplasma in cattle and Asian water buffalo in Thailand: Revealing globally distributed sequence types and regional genetic diversity
Hemotropic mycoplasmas, particularly Mycoplasma wenyonii and “Candidatus Mycoplasma haematobovis”, are increasingly recognized as emerging vector-borne pathogens of veterinary importance in cattle and buffalo. Due to their complex epidemiology and the limited understanding of their pathogenic mechanisms, hemoplasma infections in large ruminants are often underdiagnosed and underreported, especially in resource-limited settings. This study investigated the genetic diversity and phylogenetic relationships of hemotropic Mycoplasma species in blood samples collected from cattle and buffalo across seven provinces in Thailand between 2016 and 2023. A total of 422 samples (214 from cattle and 208 from buffalo) were analyzed, with 373 testing positive for hemotropic Mycoplasma DNA. Phylogenetic analyses of the 16S and 23S rRNA gene sequences revealed two distinct clades corresponding to M. wenyonii and “Ca. M. haematobovis”. Most sequences clustered with M. wenyonii strains from multiple countries, while a smaller subset grouped with “Ca. M. haematobovis” strains. Intraspecific sequence similarity among M. wenyonii 16S rRNA gene sequences ranged from 97.29% to 100%, whereas “Ca. M. haematobovis” sequences showed similarities ranging from 99.75% to 100%. Interspecific similarity between the two species ranged from 83.48% to 84.34%. For the 23S rRNA gene sequences, M. wenyonii exhibited intraspecific similarity levels ranging from 90.94% to 100%, while “Ca. M. haematobovis” sequences demonstrated 99.53% similarity; interspecific similarity for this gene ranged from 76.69% to 77.34%. We identified 11 nucleotide sequence types (ntSTs) for M. wenyonii, with ntST10 being the most widely distributed, shared among isolates from Cuba, Thailand, Japan, and Brazil. The single “Ca. M. haematobovis” sequence type detected in this study clustered within ntST7, alongside sequences from Cuba, Malaysia, Brazil, Turkey, Kyrgyzstan, and Japan. These findings highlight the substantial genetic diversity and broad geographical distribution of hemotropic Mycoplasma species in Thailand and emphasize the critical importance of molecular screening for managing the risk of pathogen transmission within livestock populations.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


