使用多序列MRI对脑癌诊断的视觉语言模型进行基准测试
Q1 Medicine
引用次数: 0
摘要
大型语言模型(llm)在各个领域的迅速采用,促使人们对其在医疗保健生态系统中的应用越来越感兴趣。特别是,视觉语言模型(VLMs)提供了生成放射学报告的潜力。本研究旨在对不同尺寸和专业领域的最先进VLMs进行综合评估,以确定其在生成脑癌患者MRI扫描放射学报告方面的有效性。我们对几个开源vlm进行了比较分析,包括Qwen2-VL、Meta-Vision 3.2、PaliGemma 2、deepseek - v2、Nvidia开源模型和医疗专用vlm。每个模型族都被评估为小、中、大的变体。由癌症患者的多序列脑MRI扫描组成的基准数据集在专家注释和委员会认证的放射科医生的指导下进行了策划。使用客观指标、推理模型(R1和01模型,根据完整性、简洁性和正确性判断AI代)和人类专家,对模型生成完整放射学报告的能力进行评估。不同大小和类型的模型性能差异很大。在大型模型中,Meta-Vision 3.2 90B的得分最高,0为70.19%,R1为68.09%。在中等类别中,Meta-Vision 11B和DeepSeek-VL-2-27B表现优异,分别获得了57.56%和53.44%的01分和51.06%和52.75%的R1分。对于较小的模型,Qwen2-VL-2B表现出最好的性能(R1: 23.88%, R1: 23.25%)。据我们所知,这是第一个评估VLMs为使用多序列MRI诊断脑癌生成综合放射学报告的研究。我们的研究结果揭示了基于模型大小和专业化的实质性性能差异,为未来发展和优化医疗vlm提供了重要指导,以支持诊断放射学工作流程。本文章由计算机程序翻译,如有差异,请以英文原文为准。
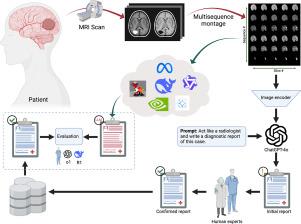
Benchmarking vision-language models for brain cancer diagnosis using multisequence MRI
The rapid adoption of Large Language Models (LLMs) in various fields has prompted growing interest in their application within the healthcare ecosystem. In particular, Vision Language Models (VLMs) offer potential for generating radiology reports. This study aims to perform a comprehensive evaluation of state-of-the-art VLMs with varying sizes and domain specializations to determine their effectiveness in generating radiology reports for MRI scans of brain cancer patients. We conducted a comparative analysis of several open-source VLMs, including Qwen2-VL, Meta-Vision 3.2, PaliGemma 2, DeepSeek-VL2, Nvidia open-source models, and medical-specific VLMs. Each model family was assessed across small, medium, and large variants. A benchmark dataset comprising multisequence brain MRI scans of cancer patients was curated with expert annotation and guidance from board-certified radiologists. The models were evaluated on their ability to generate complete radiology reports, using objective metrics, reasoning models (R1 and o1 models that judged the AI generations based on completeness, conciseness, and correctness), and human experts. Model performance varied significantly across size and type. Among large-scale models, Meta-Vision 3.2 90B achieved the highest scores with o1 of 70.19% and R1 of 68.09%. In the medium category, Meta-Vision 11B and DeepSeek-VL-2-27B outperformed others, achieving o1 scores of 57.56% and 53.44%, and R1 scores of 51.06% and 52.75%, respectively. For smaller models, Qwen2-VL-2B demonstrated the best performance (o1: 23.88%, R1: 23.25%). To our knowledge, this is the first study evaluating VLMs for generating comprehensive radiology reports for brain cancer diagnosis using multisequence MRI. Our findings reveal substantial performance differences based on model size and specialization, offering important guidance for future development and optimization of medical VLMs to support diagnostic radiology workflows.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Informatics in Medicine Unlocked
Medicine-Health Informatics
CiteScore
9.50
自引率
0.00%
发文量
282
审稿时长
39 days
期刊介绍:
Informatics in Medicine Unlocked (IMU) is an international gold open access journal covering a broad spectrum of topics within medical informatics, including (but not limited to) papers focusing on imaging, pathology, teledermatology, public health, ophthalmological, nursing and translational medicine informatics. The full papers that are published in the journal are accessible to all who visit the website.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


