pq3 -喹啉衍生物作为选择性COX-2抑制剂的多尺度研究:合成、结构分析、DFT、MD模拟和硅/体外评价。
IF 4.7
2区 医学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
喹诺喹啉由于其结构的多功能性和广泛的药理潜力,已成为药物发现,特别是抗炎药开发中有前途的支架。其灵活的化学框架允许进行多种修饰,从而能够设计出针对与癌症进展有关的关键分子途径的衍生物。以3-苯基喹啉-2(1H)- 1(1)为原料,与1-溴烷(2a-f)和四丁基溴化铵(BTBA)在DMF中以较好的收率合成了标题化合物3-苯基喹啉-2(1H)- 1 (PQ3a-f)。对得到的粗产物进行了再结晶,并通过1HNMR、13CNMR和LC-MS等光谱技术对其进行了鉴定,通过单晶x射线衍射(XRD)研究证实了PQ3a的三维晶体结构。Hirshfeld表面分析证实了C-H…O分子间的相互作用。PQ3a的二维指纹图谱显示,HH接触对整个Hirshfeld表面积的贡献最大(72.7%)。三维能量框架计算表明,大部分色散能量超过其他能量。通过DFT计算得到PQ3a在B3LYP/6-31 + G(d,p)基组下的分子性质。HOMO-LUMO之间的能隙为3.81 eV,表明其具有很强的生物活性。在所合成的6个PQ3a-f衍生物中,PQ3a对COX-2的抑制作用最强(IC50 = 10.24 μM),对COX-1的抑制作用中等(IC50 = 31.67 μM),选择性指数(SI)为3.09,表明PQ3a-f衍生物对COX-2具有选择性。PQ3f对COX-2 (IC50 = 25.54 μM)和COX-1 (IC50 = 46.45 μM)具有中度抑制作用,SI为1.82。相比之下,其余衍生物(PQ3b-d)与参比药物塞来昔布相比表现出弱或非选择性的COX抑制作用(COX-2 IC50 = 0.54 μM; SI = 21.15)。对接结果显示,与Ser530和Leu531的关键相互作用支持了强的结合亲和力,特别是与COX-2的结合。分子动力学模拟(MDs)证实了PQ3a-COX-2复合物在500 ns内的稳定性,突出了持续的疏水相互作用和与Ser530的稳定氢键。这些发现表明PQ3a是一种很有前途的选择性COX-2抑制剂,用于抗炎治疗。本文章由计算机程序翻译,如有差异,请以英文原文为准。
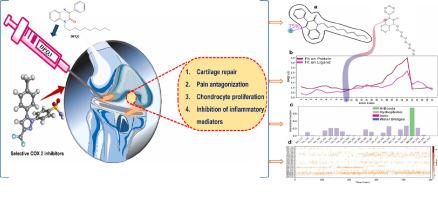
Multi-scale investigation of PQ3-quinoxaline derivatives as selective COX-2 inhibitors: synthesis, structural analysis, DFT, MD simulations, and in silico/in vitro evaluation
Quinoxaline has emerged as a promising scaffold in drug discovery, particularly in the development of anti-inflammatory agents, due to its structural versatility and broad pharmacological potential. Its flexible chemical framework permits diverse modifications, enabling the design of derivatives that can target critical molecular pathways implicated in cancer progression. The title compounds of 3-phenylquinoxalin-2(1H)-one (PQ3a-f), were synthesized in good yields from 3-phenylquinoxalin-2(1H)-one (1) in DMF, with 1-bromoalkanes (2a-f), and tetrabutylammonium bromide (BTBA). The crude products obtained have been recrystallized and elucidated by spectroscopic techniques (1HNMR, 13CNMR and LC-MS), the 3D of PQ3a crystal structure was confirmed by single crystal X-ray diffraction (XRD) studies. Hirshfeld surface analysis confirmed the C-H…O intermolecular interactions. 2D fingerprint plot of PQ3a shows that the major contribution to the overall Hirshfeld surface area is from H![]() H (72.7 %) contacts. 3D energy-frameworks calculations shows that the majority of dispersion energy over the other energies. DFT calculations are performed to know the PQ3a molecular properties at B3LYP/6–31 + G(d,p) basis sets. The energy gap between the HOMO-LUMO is found to be 3.81 eV, indicating the nature of potent biologically active molecule. Among the six synthesized PQ3a-f derivatives, PQ3a exhibited the most potent COX-2 inhibition (IC50 = 10.24 μM) with moderate COX-1 inhibition (IC50 = 31.67 μM), yielding a selectivity index (SI) of 3.09, indicative of COX-2 selectivity. PQ3f showed moderate COX-2 inhibition (IC50 = 25.54 μM) and COX-1 inhibition (IC50 = 46.45 μM), with an SI of 1.82. In contrast, the remaining derivatives (PQ3b-d) demonstrated weak or non-selective COX inhibition compared to the reference drug celecoxib (COX-2 IC50 = 0.54 μM; SI = 21.15). Docking results revealed strong binding affinity, particularly with COX-2, supported by key interactions with Ser530 and Leu531. Molecular dynamics simulations (MDs) confirmed the stability of the PQ3a-COX-2 complex over 500 ns, highlighting sustained hydrophobic interactions and a stable hydrogen bond with Ser530. These findings suggest PQ3a as a promising selective COX-2 inhibitor for anti-inflammatory therapy.
H (72.7 %) contacts. 3D energy-frameworks calculations shows that the majority of dispersion energy over the other energies. DFT calculations are performed to know the PQ3a molecular properties at B3LYP/6–31 + G(d,p) basis sets. The energy gap between the HOMO-LUMO is found to be 3.81 eV, indicating the nature of potent biologically active molecule. Among the six synthesized PQ3a-f derivatives, PQ3a exhibited the most potent COX-2 inhibition (IC50 = 10.24 μM) with moderate COX-1 inhibition (IC50 = 31.67 μM), yielding a selectivity index (SI) of 3.09, indicative of COX-2 selectivity. PQ3f showed moderate COX-2 inhibition (IC50 = 25.54 μM) and COX-1 inhibition (IC50 = 46.45 μM), with an SI of 1.82. In contrast, the remaining derivatives (PQ3b-d) demonstrated weak or non-selective COX inhibition compared to the reference drug celecoxib (COX-2 IC50 = 0.54 μM; SI = 21.15). Docking results revealed strong binding affinity, particularly with COX-2, supported by key interactions with Ser530 and Leu531. Molecular dynamics simulations (MDs) confirmed the stability of the PQ3a-COX-2 complex over 500 ns, highlighting sustained hydrophobic interactions and a stable hydrogen bond with Ser530. These findings suggest PQ3a as a promising selective COX-2 inhibitor for anti-inflammatory therapy.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Bioorganic Chemistry
生物-生化与分子生物学
CiteScore
9.70
自引率
3.90%
发文量
679
审稿时长
31 days
期刊介绍:
Bioorganic Chemistry publishes research that addresses biological questions at the molecular level, using organic chemistry and principles of physical organic chemistry. The scope of the journal covers a range of topics at the organic chemistry-biology interface, including: enzyme catalysis, biotransformation and enzyme inhibition; nucleic acids chemistry; medicinal chemistry; natural product chemistry, natural product synthesis and natural product biosynthesis; antimicrobial agents; lipid and peptide chemistry; biophysical chemistry; biological probes; bio-orthogonal chemistry and biomimetic chemistry.
For manuscripts dealing with synthetic bioactive compounds, the Journal requires that the molecular target of the compounds described must be known, and must be demonstrated experimentally in the manuscript. For studies involving natural products, if the molecular target is unknown, some data beyond simple cell-based toxicity studies to provide insight into the mechanism of action is required. Studies supported by molecular docking are welcome, but must be supported by experimental data. The Journal does not consider manuscripts that are purely theoretical or computational in nature.
The Journal publishes regular articles, short communications and reviews. Reviews are normally invited by Editors or Editorial Board members. Authors of unsolicited reviews should first contact an Editor or Editorial Board member to determine whether the proposed article is within the scope of the Journal.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


