基于隐私保护分子生成的脑启发联合扩散变压器
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
目的:生成式药物发现受到数据隐私挑战和SOTA模型巨大计算成本的阻碍。为了克服这些障碍,我们开发了一种保护隐私和资源高效的框架——脑启发联合扩散强化(BiFDR)。方法:BiFDR集成三个协同模块。受神经启发的联邦协调器(NeuroFed)通过受突触可塑性启发的原则编排安全协作,将服务器端修剪与客户端低秩适应(LoRA)和稀疏异步更新相结合。基于变压器的扩散发生器(TransFuse)利用注意力机制在压缩的潜在空间中有效地产生化学有效分子。最后,一个强化学习代理(T-JORM)将生成过程转向新的2D和3D分子结构,由一个多方面的、基于谷本的奖励函数指导。结果:以基线模型为基准,BiFDR将药物相似性定量估计提高了13.7%,分子水平结构信息评分提高了5.7%,分子相互作用分析指数提高了52.3%。该框架还增强了综合可行性,综合可达性得分降低了9.5%。关键的是,BiFDR大大加强了数据隐私,实现了互信息度量减少43.6%。结论:BiFDR为生成性药物发现建立了一个有效和高效的范式。它始终如一地产生具有优异的药物相似性、结构新颖性和相互作用潜力的分子。通过确保合成可及性,同时严格保护隐私并最大限度地减少计算开销,BiFDR为现代协作药物开发管道提供了可行且可扩展的解决方案。本文章由计算机程序翻译,如有差异,请以英文原文为准。
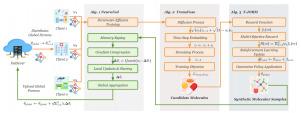
BiFDR: Brain-Inspired Federated Diffusion Transformer with Reinforcement for privacy-preserving molecular generation
Objective:
Generative drug discovery is hampered by challenges in data privacy and the immense computational cost of SOTA models. To surmount these barriers, we developed Brain-Inspired Federated Diffusion with Reinforcement (BiFDR), a privacy-preserving and resource-efficient framework.
Methods:
BiFDR integrates three synergistic modules. A Neuro-inspired Federated Coordinator (NeuroFed) orchestrates secure collaboration via synaptic plasticity-inspired principles, combining server-side pruning with client-side Low-Rank Adaptation (LoRA) and sparse asynchronous updates. A Transformer-based diffusion generator (TransFuse) efficiently creates chemically valid molecules in a compressed latent space using attention mechanisms. Finally, a reinforcement learning agent (T-JORM) steers the generative process towards novel 2D and 3D molecular structures, guided by a multi-faceted, Tanimoto-based reward function.
Results:
Benchmarked against baseline models, BiFDR improving the Quantitative Estimate of Drug-likeness by 13.7%, the Molecular-level Structural Information Score by 5.7%, and the Molecular Interaction Analysis Index by 52.3%. The framework also enhanced synthetic feasibility, reflected by a 9.5% reduction in the Synthetic Accessibility Score. Critically, BiFDR substantially strengthened data privacy, achieving a 43.6% reduction in the mutual information metric.
Conclusion:
BiFDR establishes an effective and efficient paradigm for generative drug discovery. It consistently produces molecules with superior drug-likeness, structural novelty, and interaction potential. By ensuring synthetic accessibility while rigorously preserving privacy and minimizing computational overhead, BiFDR presents a viable and scalable solution for modern, collaborative drug development pipelines.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


