石田小仓鼠(半翅目:仓鼠科)吡虫啉抗性研究:RNA-Seq和RT-qPCR功能验证
IF 2.2
2区 生物学
Q4 BIOCHEMISTRY & MOLECULAR BIOLOGY
Comparative Biochemistry and Physiology D-Genomics & Proteomics
Pub Date : 2025-09-08
DOI:10.1016/j.cbd.2025.101630
引用次数: 0
摘要
杀虫剂抗药性的迅速发展是农业害虫治理面临的日益严峻的挑战。棉花和其他作物的主要吸液害虫——小圆蝽(Amrasca biguttula biguttula)通过取食和注射毒素造成严重损害,同时对吡虫啉等多种杀虫剂具有抗性。了解小圆蝽抗性的遗传因素和机制对有效防治具有重要意义。本研究利用Illumina HiSeq测序生成转录组组装,并分析吡虫啉对差异基因表达的响应。使用DESeq2进行差异表达分析,鉴定出177个显著表达的转录本,其中67个上调,110个下调。为了验证这些发现,我们在不同时间点(6小时、12小时和24小时)相对于0小时(对照组)测试了LC50浓度(24.91 ppm)吡虫啉暴露后选定基因的基因表达。解毒相关基因如细胞色素P450单加氧酶(CYP303A1、CYP4DE1、CYP3184-fragment1和CYP3939A2)、三种羧酸酯酶(CE-1、CE-2和CE-3)和一种ABC转运蛋白(ABC)的显著上调,突出了其在吡虫啉耐药中的潜在作用。本研究首次探索了异毛小仓鼠对吡虫啉抗性的分子机制,为改进当前的害虫防治策略提供了有价值的见解。本文章由计算机程序翻译,如有差异,请以英文原文为准。
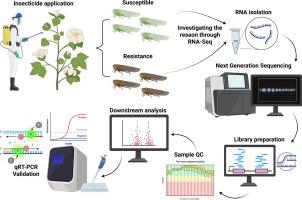
Investigating imidacloprid resistance in Amrasca biguttula biguttula (Ishida) (Hemiptera: Cicadellidae): Insights from RNA-Seq and functional validation using RT-qPCR
The increasing challenge in agricultural insect pest management is the rapid development of insecticide resistance. Amrasca biguttula biguttula, a significant sap-sucking pest of cotton and other crops, causes severe damage through feeding and toxin injection while exhibiting resistance to multiple insecticides, including Imidacloprid. Understanding the genetic factors and mechanisms driving resistance in A. biguttula biguttula is crucial for effective pest control. This study utilized Illumina HiSeq sequencing to generate a transcriptome assembly and analyze differential gene expression in response to Imidacloprid. Differential expression analysis using DESeq2 identified 177 significantly expressed transcripts, with 67 upregulated and 110 downregulated. To validate these findings, the gene expression of selected genes in response to Imidacloprid exposure at LC50 concentration (24.91 ppm) was tested at different time points (6 h, 12 h, and 24 h) relative to 0 h (control). The significant upregulation of detoxification-related genes such as cytochrome P450 monooxygenase (CYP303A1, CYP4DE1, CYP3184-fragment1 and CYP3939A2), three carboxylesterases (CE-1, CE-2, and CE-3), and one ABC transporter (ABC), highlights its potential role in Imidacloprid resistance. This study is the first to explore the molecular mechanisms underlying Imidacloprid resistance in A. biguttula biguttula and offers valuable insights for improving current pest management strategies.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
CiteScore
5.10
自引率
3.30%
发文量
69
审稿时长
33 days
期刊介绍:
Comparative Biochemistry & Physiology (CBP) publishes papers in comparative, environmental and evolutionary physiology.
Part D: Genomics and Proteomics (CBPD), focuses on “omics” approaches to physiology, including comparative and functional genomics, metagenomics, transcriptomics, proteomics, metabolomics, and lipidomics. Most studies employ “omics” and/or system biology to test specific hypotheses about molecular and biochemical mechanisms underlying physiological responses to the environment. We encourage papers that address fundamental questions in comparative physiology and biochemistry rather than studies with a focus that is purely technical, methodological or descriptive in nature.

 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


