抗毒素RNA假结在调节毒素活性和毒素-抗毒素RNP复合物组装中的作用
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
毒素-抗毒素(TA)系统是细菌在压力下赋予生存优势的防御系统。TA系统包括一个毒素和一个抗毒素基因,通常作为操纵子存在于染色体或细菌的质粒上。在III型毒素TA系统中,毒素基因编码一种蛋白毒素(ToxN),这是一种序列特异性核糖核酸内切酶,而抗毒素基因编码一种RNA抗毒素(ToxI),通过形成一个封闭的环状TA RNP复合物来中和毒素。在TA组装中,抗毒素RNA采用复杂的三级结构,包括一个中央保守的假结,两侧是毒素结合的5 ‘和3 ’单链区域。在这项研究中,我们已经证明,在大肠杆菌中,毒素RNP复合物的一个封闭的环状组装是完全抑制毒素的必要条件。我们已经探索了抗毒素假结内的三级接触,这是大肠杆菌毒素抑制所必需的。此外,我们通过体外生物物理和生化实验研究了几种ToxI突变体对抗毒素RNA稳定性、结构和TA复合物组装的影响。我们已经证明ToxI突变体采用不同于功能性ToxI重复序列的结构。在改变的构象中,ToxI突变体能够结合毒素,但不能组装成封闭的组装体,导致毒素的不完全抑制。我们的研究结果表明,假结中细微的核苷酸变化可以破坏抗毒素介导的毒素中和,强调其在TA复合物组装中的作用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
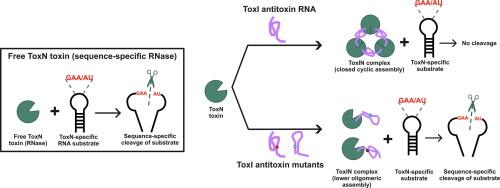
Role of Antitoxin RNA Pseudoknot in Regulating Toxin Activity and Toxin-antitoxin RNP Complex Assembly
Toxin-antitoxin (TA) systems are bacterial defense systems that confer survival advantages under stress. TA systems comprise a toxin and an antitoxin gene, usually present as operon on chromosomes or on plasmids in bacteria. In type III ToxIN TA systems toxin gene encodes a protein toxin (ToxN) which is a sequence-specific endoribonuclease and antitoxin gene encodes an RNA antitoxin (ToxI) that neutralizes toxin by forming a closed-cyclic TA RNP complex. In TA assemblies, antitoxin RNA adopts a complex tertiary structure comprising of a central conserved pseudoknot flanked by toxin-binding 5′ and 3′ single-stranded regions. In this study, we have shown that a closed, cyclic assembly of ToxIN RNP complex is required for the complete ToxN inhibition in E. coli. We have probed tertiary contacts within the antitoxin pseudoknot that are essential for toxin inhibition in E. coli. Furthermore, we investigated the impact of several ToxI mutants on antitoxin RNA stability, structure, and TA complex assembly using in vitro biophysical and biochemical experiments. We have shown that ToxI mutants adopt structures different from the functional ToxI repeat. In altered conformations, ToxI mutants were able to bind the toxin but were unable to assemble into closed assemblies, resulting in incomplete inhibition of the toxin. Our findings showed that subtle nucleotide changes in the pseudoknot can disrupt antitoxin-mediated toxin neutralization, emphasizing its role in TA complex assembly.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


