PCGMMF:基于增强多模态特征融合的乳腺癌预后复发转移风险预测方法。
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
背景:乳腺癌是一种高发病率和高死亡率的异质性疾病。尽管有各种治疗方法,但仍有相当一部分患者面临复发或转移的高概率,严重影响其生存状态。基于单模态数据和机器学习算法的传统预后方法往往无法充分捕捉乳腺癌复杂的生物学关系和异质性特征,导致预后表现不佳。因此,迫切需要一种更准确有效的预测乳腺癌复发转移风险的预后方法。方法:在本研究中,我们提出了一种称为PCGMMF的新方法用于乳腺癌预后分析。该方法通过多模态融合整合组织病理图像、临床数据、基因表达数据和DNA甲基化数据。我们利用基于迁移学习的预训练视觉- lstm模型从组织病理学图像中提取特征。此外,我们设计了一个综合的特征选择策略,包括支持向量机(SVM)、Mantel测试和相关分析,从基因表达数据和DNA甲基化数据中过滤特征。此外,为了解决乳腺癌的高度异质性和多模态特征的独立性和交叉性,我们提出了一个基于双向关注和自关注的增强多模态特征融合模块BSAMF。结果:通过一系列实验,对PCGMMF的性能进行了评价。在预测乳腺癌复发转移风险时,PCGMMF的准确率为0.903,AUC值为0.924,优于其他最先进的方法。此外,我们还提供了组织病理学图像中高度显著区域的可解释性分析,可作为临床实践的参考。结论:PCGMMF通过有效整合多模态数据和利用先进的深度学习技术,为乳腺癌预后分析提供了一个强大而创新的解决方案。可有效进行乳腺癌预后分析,为个性化精准治疗及临床实践提供重要参考。本文章由计算机程序翻译,如有差异,请以英文原文为准。
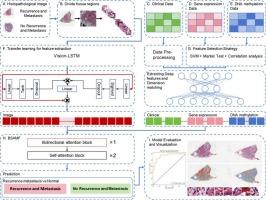
PCGMMF: a prediction method for breast cancer prognostic recurrence and metastasis risk based on enhanced multimodal feature fusion
Background
Breast cancer is a highly heterogeneous disease with high morbidity and mortality rates. Despite the availability of various treatments, a significant number of patients still face a high probability of recurrence or metastasis, which severely impacts their survival status. Traditional prognostic methods based on single-modality data and machine learning algorithms often fail to adequately capture the complex biological relationships and heterogeneous characteristics of breast cancer, leading to suboptimal prognostic performance. Therefore, there is an urgent need for a more accurate and effective method to predict the risk of recurrence and metastasis in breast cancer prognosis.
Methods
In this study, we propose a novel method termed PCGMMF for breast cancer prognostic analysis. This method integrates histopathological images, clinical data, gene expression data, and DNA methylation data through multimodal fusion. We leverage a pre-trained Vision-LSTM model based on transfer learning to extract features from histopathological images. Additionally, we design a comprehensive feature selection strategy that includes support vector machine (SVM), Mantel test, and correlation analysis to filter features from gene expression data and DNA methylation data. Furthermore, to address the high heterogeneity of breast cancer and the independence and intersectionality of multimodal features, we propose a bidirectional attention and self-attention based enhanced multimodal feature fusion module called BSAMF.
Results
Through a series of experiments, we evaluate the performance of PCGMMF. When predicting the recurrence and metastasis risk of breast cancer prognosis, PCGMMF achieves an accuracy of 0.903 and an AUC value of 0.924, outperforming other state-of-the-art methods. Furthermore, we provide an interpretability analysis of highly significant regions from histopathological images, which can serve as a reference for clinical practice.
Conclusion
PCGMMF offers a robust and innovative solution for breast cancer prognostic analysis by effectively integrating multimodal data and utilizing advanced deep learning techniques. It can effectively conduct breast cancer prognostic analysis and provide significant references for personalized precision treatment and clinical practice.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


