与血浆基准相协调的人血浆代谢物注释
IF 20.8
1区 医学
Q1 ENDOCRINOLOGY & METABOLISM
引用次数: 0
摘要
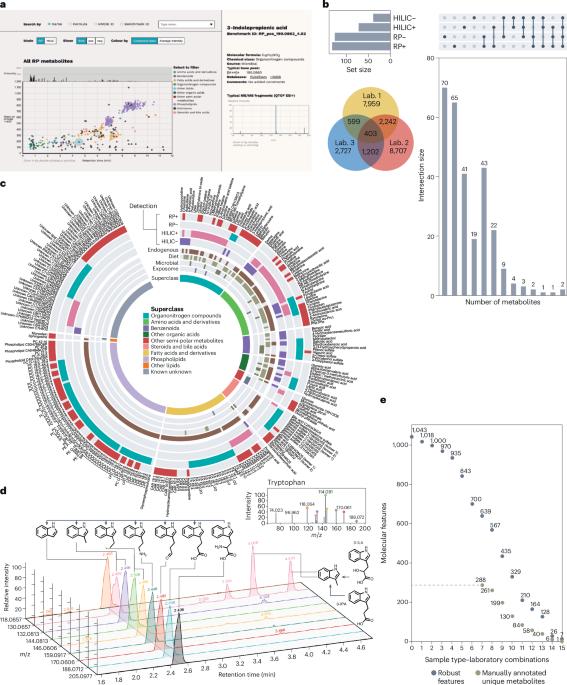
人类血浆代谢组已被广泛表征:人类代谢组数据库(HMDB)1 5.0版目前包含37,229个人类血液中报告的代谢物条目。尽管现代LC-MS平台的分析覆盖范围能够检测和鉴定1,000-2,000种血浆代谢物,但已知和定期检测的血浆代谢物的数量相当少:健康人群中144种血浆代谢物的参考值2和美国国家标准与技术研究所(NIST)参考血浆样品3中的588种脂质已被报道。血浆代谢组也表现出巨大的个体间和个体内部差异,这可以通过微生物组、饮食习惯和遗传的差异来解释。持续报告人类血浆代谢组的挑战来自于非靶向LC-MS方法的实验室间差异以及代谢物本身注释的不同实践和能力5。为了构建Plasma Benchmark,三个参与实验室以四种分析模式分析了相同的内部池等离子体和NIST1950人类参考等离子体6,包括正负电离模式下的反相和亲水相互作用色谱,然后在MS-DIAL7中生成数据矩阵。我们应用了一套基于信噪比、相对标准差、样本空白比、MS/MS数据采集和预测分子式的纳入标准,将检测到的分子特征数量缩小到639个最可能代表实际血浆代谢物的强大分子特征。近88%的检测到的分子特征不符合信噪比和样本空白比标准,这可能是由于LC-MS数据中固有的噪声和污染物,也可能是由于实验室中许多分子特征的检测具有很高的可变性8(图1b)。人工筛选得到288个独特的代谢物,而其余的分子特征被归类为冗余分子特征和源内片段。包含四种分析模式对于代谢物的广泛覆盖至关重要,因为大多数代谢物仅在一种模式下检测到(图1b)。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Harmonizing human plasma metabolite annotation with Plasma Benchmark
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊
Nature metabolism
ENDOCRINOLOGY & METABOLISM-
CiteScore
27.50
自引率
2.40%
发文量
170
期刊介绍:
Nature Metabolism is a peer-reviewed scientific journal that covers a broad range of topics in metabolism research. It aims to advance the understanding of metabolic and homeostatic processes at a cellular and physiological level. The journal publishes research from various fields, including fundamental cell biology, basic biomedical and translational research, and integrative physiology. It focuses on how cellular metabolism affects cellular function, the physiology and homeostasis of organs and tissues, and the regulation of organismal energy homeostasis. It also investigates the molecular pathophysiology of metabolic diseases such as diabetes and obesity, as well as their treatment. Nature Metabolism follows the standards of other Nature-branded journals, with a dedicated team of professional editors, rigorous peer-review process, high standards of copy-editing and production, swift publication, and editorial independence. The journal has a high impact factor, has a certain influence in the international area, and is deeply concerned and cited by the majority of scholars.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


