通过丰富的验证和有针对性的抽样来克服数据挑战,以测量电子健康记录中的整个人的健康。
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
目的:适应负荷指数(ALI)是一个由10个成分组成的整体人体健康指标,它反映了健康功能背后的多个相互关联的生理调节系统。来自电子健康记录(EHR)的数据为在学习卫生系统中实施ALI提供了巨大的机会;然而,这些数据容易丢失和错误。验证(例如,通过图表审查)可以提供更高质量的数据,但实际上,只有一小部分患者的数据可以被验证,而且大多数方案不能恢复丢失的数据。方法:从一个广泛的学习型卫生系统的电子病历中选取1000名患者作为代表性样本(其中100人可以被验证),我们提出了设计、实施和分析ALI和医疗保健利用的统计有效和稳健研究的方法。采用半参数最大似然估计,我们稳健地将所有可用的患者信息纳入统计模型。使用有针对性的设计策略,我们检查了选择最具信息性的患者进行验证的方法。结合临床专业知识,我们设计了一种新的验证方案,以提高电子病历数据的质量和完整性。结果:图表审核发现的错误很少(99%与源文档匹配),并通过患者图表中的辅助信息恢复了一些缺失的数据。平均而言,验证将每位患者非缺失ALI成分的数量从6个增加到7个。通过基于初步数据的模拟,残差抽样被确定为完成我们验证研究的最具信息性的策略。结合验证数据,统计模型表明,整体健康状况较差(ALI较高)与参与医疗保健系统的几率较高相关,并根据年龄进行调整。结论:采用丰富的方案进行有针对性的验证,可以保证电子病历数据的质量,提高数据的完整性。我们验证研究的结果被纳入分析,因为我们将ALI作为可扩展的全人健康测量来预测学习健康系统中的医疗保健利用。本文章由计算机程序翻译,如有差异,请以英文原文为准。
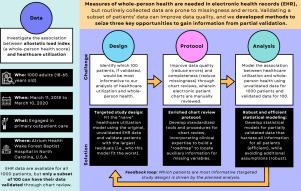
Overcoming data challenges through enriched validation and targeted sampling to measure whole-person health in electronic health records
Objective:
The allostatic load index (ALI) is a 10-component composite measure of whole-person health, which reflects the multiple interrelated physiological regulatory systems that underlie healthy functioning. Data from electronic health records (EHR) present a huge opportunity to operationalize the ALI in learning health systems; however, these data are prone to missingness and errors. Validation (e.g., through chart reviews) can provide better-quality data, but realistically, only a subset of patients’ data can be validated, and most protocols do not recover missing data.
Methods:
Using a representative sample of 1000 patients from the EHR at an extensive learning health system (100 of whom could be validated), we propose methods to design, conduct, and analyze statistically efficient and robust studies of ALI and healthcare utilization. Employing semiparametric maximum likelihood estimation, we robustly incorporate all available patient information into statistical models. Using targeted design strategies, we examine ways to select the most informative patients for validation. Incorporating clinical expertise, we devise a novel validation protocol to promote EHR data quality and completeness.
Results:
Chart reviews uncovered few errors (99% matched source documents) and recovered some missing data through auxiliary information in patients’ charts. On average, validation increased the number of non-missing ALI components per patient from 6 to 7. Through simulations based on preliminary data, residual sampling was identified as the most informative strategy for completing our validation study. Incorporating validation data, statistical models indicated that worse whole-person health (higher ALI) was associated with higher odds of engaging in the healthcare system, adjusting for age.
Conclusion:
Targeted validation with an enriched protocol can ensure the quality and promote the completeness of EHR data. Findings from our validation study were incorporated into analyses as we operationalize the ALI as a scalable whole-person health measure that predicts healthcare utilization in the learning health system.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


