评价AlphaFold3对荧光蛋白翻译后修饰、寡聚物组装和可淬灭金属结合的预测
IF 3
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
绿色荧光蛋白(gfp)是一种光学标记物,广泛用于分子和细胞生物学研究,以跟踪生物分子的位置和功能。阐明它们的结构将有助于这些荧光蛋白的进一步工程设计,以提高其性能。AlphaFold3 (AF3)是最近开发的一种预测工具,与其他预测工具相比,它具有更高的准确性,特别是在预测蛋白质与配体相互作用时与最先进的对接工具相比。然而,利用AF3分析FPs结构的研究尚未充分开展。为了确定利用AF3预测FP的准确性,我们分析了FP的发色团形成、寡聚态和金属离子结合,并与实验测定的FP进行了比较。AF3不能通过翻译后修饰产生包含双环结构的发色团。此外,FPs的低聚体组装形成与实验低聚体FPs相似;然而,FP单体之间的残留相互作用的细节是不同的。使用AF3与FPs进行可淬灭金属离子对接,发现了类似的金属结合位点;然而,AF3与实验结构之间的金属配位存在显著差异。这些结果揭示了使用AF3治疗FPs的潜力和局限性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
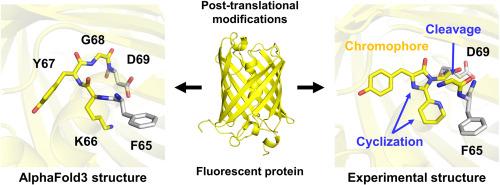
Evaluation of AlphaFold3 prediction for post-translational modification, oligomeric assembly, and quenchable metal binding of fluorescent proteins
Green fluorescent proteins (GFPs) are optical markers that are widely used in molecular and cell biology studies to track the location and function of biomolecules. Elucidating their structures will facilitate further engineering of these fluorescent proteins (FPs) to enhance their properties. AlphaFold3 (AF3) is a recently developed prediction tool that exhibits higher accuracy compared with other prediction tools, particularly in predicting protein–ligand interactions with state-of-the-art docking tools. However, studies on the use of AF3 to analyze the structure of FPs have not been fully conducted. To determine the accuracy of FP prediction using AF3, chromophore formation, oligomeric states, and metal ion binding of FPs were analyzed and compared with those of experimentally determined FPs. AF3 could not generate a chromophore comprising the two-ring structure by post-translational modification. Moreover, the oligomeric assembly formation of the FPs was similar to that of experimental oligomeric FPs; however, the detailed residue interactions between FP monomers were different. Quenchable metal ion docking to FPs using AF3 revealed a similar metal-binding site; however, metal coordination was significantly different between AF3 and the experimental structure. These results provide insight into the potential and limitations of using AF3 for FPs.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


