染色质结合调节由朊病毒样结构域形成的凝聚物的相行为和形态。
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
许多转录因子(tf)含有内在无序区(IDRs),并被认为在细胞核中形成生物分子凝聚体。这些蛋白可以被定义为阻滞共聚物,idr驱动同型和异型蛋白-蛋白相互作用,dna结合域(DBD)介导与染色质的异型相互作用。虽然体外研究主要报道了在没有染色质的情况下微米尺度的球形凝聚体,但活细胞中的TF凝聚体表现出截然不同的行为——采用多种纳米尺度的非球形形态,并表现出亚扩散动力学。在这里,我们展示了这种独特的相行为可以由tf -染色质相互作用产生,使用具有可调IDR-DBD结构的工程融合蛋白。具体来说,我们将哺乳动物SWI/SNF复合物中SS18亚基的朊病毒样结构域(PLD)(已知驱动同型相分离的结构域)融合到先锋因子FOXA1的DBD上。过表达的SS18PLD在细胞中形成较大的球形凝析物,与FOXA1DBD融合后,凝析物重新定位于染色质,呈现非球形形态,并表现出染色质湿润行为。dbd -染色质结合的破坏使凝析物形态向混合或球形状态转变,这表明染色质亲和力是凝析物粗化和空间组织的关键调节因子。粗粒度模拟总结了这些观察结果,揭示了PLD-PLD和DBD-DNA相互作用之间的精细平衡,共同决定了凝析液的动力学和结构。总之,我们的研究结果表明,染色质结合是体内转录凝聚行为的关键调节剂。本文章由计算机程序翻译,如有差异,请以英文原文为准。
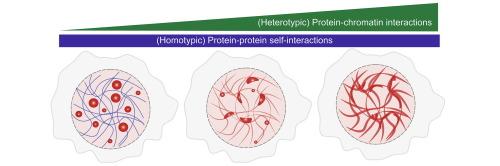
Chromatin Binding Regulates Phase Behavior and Morphology of Condensates Formed by Prion-like Domains
Many transcription factors (TFs) contain intrinsically disordered regions (IDRs) and are thought to form biomolecular condensates in the cell nucleus. These proteins can be conceptualized as block co-polymers, with the IDRs driving both homotypic and heterotypic protein–protein interactions and the DNA-binding domain (DBD) mediating heterotypic interactions with chromatin. While in vitro studies have predominantly reported micron-scale, spherical condensates in the absence of chromatin, TF condensates in live cells exhibit strikingly different behavior, adopting diverse, nanoscale, often aspherical morphologies and displaying sub-diffusive dynamics. Here, using engineered fusion proteins with tunable IDR-DBD architectures, we show that this distinct phase behavior can arise from TF-chromatin interactions. Specifically, we fused the prion-like domain (PLD) of the SS18 subunit from the mammalian SWI/SNF complex, a domain known to drive homotypic phase separation, to the DBD of the pioneer factor FOXA1. While SS18PLD on overexpression forms large, spherical condensates in cells, its fusion with FOXA1DBD leads to condensates that re-localize to chromatin, adopt aspherical morphologies, and exhibit chromatin-wetting behavior. Disruption of DBD-chromatin binding shifts condensate morphology toward a mixed or spherical state, implicating chromatin affinity as a key regulator of condensate coarsening and spatial organization. Coarse-grained simulations recapitulate these observations, revealing a finely balanced interplay between PLD-PLD and DBD-DNA interactions that collectively determine condensate dynamics and structure. Together, our findings demonstrate that chromatin binding is a critical modulator of transcriptional condensate behavior in vivo.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


