McsB通过外周精氨酸磷酸化调节CtsR的热感。
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
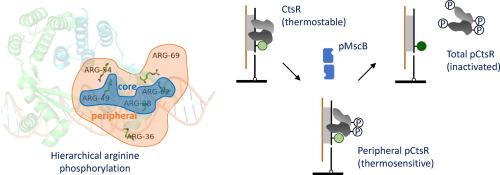
当细胞感知环境中的温度升高时,细菌的主转录抑制因子CtsR被精氨酸激酶McsB磷酸化并失活,从而启动热休克基因的表达。在这里,我们利用基于光异构相关荧光增强(或先前的蛋白质诱导荧光增强,PIFE)效应的荧光强度位移测定(FISA)来实时监测DNA-CtsR-McsB相互作用。我们的单分子分析表明,CtsR与同源DNA结合迅速而稳定,McsB能够在靶DNA的原位与CtsR短暂相互作用。我们通过单分子实时结合测定McsB与dna结合的CtsR的结合动力学,kon和koff分别为0.75 μM-1 s-1和0.34 s-1。这种与McsB的相互作用不会将CtsR从DNA中去除,但会降低CtsR解离的温度阈值并改变其热感行为。质谱分析、突变分析和结构模拟结果共同表明,CtsR上几个外周精氨酸残基的磷酸化降低了CtsR- dna复合物的结合能,这可能是这种作用的分子机制。综上所述,这些结果提供了McsB如何调节CtsR- dna相互作用的见解,并强调了CtsR外周精氨酸残基在细菌热休克反应中的功能重要性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
McsB Regulates CtsR Thermosensing Through Peripheral Arginine Phosphorylation
When cells sense an elevated temperature in the environment, the bacterial master transcription repressor CtsR becomes phosphorylated and inactivated by the arginine kinase McsB to initiate the expression of heat-shock genes. Here, we utilize a fluorescence intensity shift assay (FISA) based on the photoisomerisation-related fluorescence enhancement (or previously protein-induced fluorescence enhancement, PIFE) effect to monitor the DNA-CtsR-McsB interactions in real time. Our single-molecule analysis reveals that CtsR binds rapidly and stably to the cognate DNA, and that McsB is able to transiently interact with CtsR in situ of the target DNA. We determine the binding kinetics between McsB and the DNA-bound CtsR by single-molecule real-time binding assays, with kon and koff of 0.75 μM−1 s−1 and 0.34 s−1, respectively. This interaction with McsB does not remove CtsR from the DNA, but lowers the temperature threshold for CtsR dissociation and alters its thermosensing behavior. Mass spectrometry, mutational analysis and structural simulation results together suggest that the phosphorylation of several peripheral arginine residues on CtsR, which reduces the binding energy of the CtsR-DNA complex, underlies a plausible molecular mechanism for this effect. Taken together, these results provide insights into how McsB regulates the CtsR-DNA interaction and highlight the functional importance of CtsR peripheral arginine residues in the bacterial heat-shock response.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


