生物医学命名实体识别大型语言模型的资源高效指令调优
IF 4.5
2区 医学
Q2 COMPUTER SCIENCE, INTERDISCIPLINARY APPLICATIONS
引用次数: 0
摘要
目的:大型语言模型(LLMs)在自然语言处理(NLP)任务中表现出显著的有效性,其中生物医学命名实体识别(BioNER)的微调受到了重要的研究关注。然而,与微调大规模模型相关的大量计算需求限制了它们的开发和部署。因此,本研究探讨了参数高效微调(PEFT)技术,以在有限的计算资源下优化BioNER的llm。通过利用这些方法,在保持域内泛化能力的同时保持了具有竞争力的模型性能。方法:本研究采用PEFT方法QLoRA对开源Llama3.1模型进行微调,开发专门针对BioNER任务设计的NERLlama3.1模型。首先,利用NCBI-disease、BC5CDR-chem、BC2GM-gene等BioNER数据集创建LLM指令调优数据集。接下来,在单个16GB内存GPU上,使用QLoRA方法对Llama3.1-8B模型进行微调。此外,在推理阶段,我们引入了一种提示工程技术,称为自一致性NER提示(SCNP)。这种方法利用llm产生的输出的多样性来显著提高NER性能。最后,我们还开发了一个多任务BioNER-capable模型NERLlama3.1-MT,以研究微调llm在解决多任务BioNER场景中的能力。结果:NERLlama3.1模型在ncbi -疾病、bc5cdr -化学和bg2gm -基因数据集上的f1得分分别为0.8977、0.9402和0.8530。此外,当对以前未见过的数据集进行评估时,BC5CDR-disease的f1得分为0.6867,NLM-chemical的得分为0.6800,NLM-gene的得分为0.8378。这些结果表明,与基于bert的模型相比,NERLlama3.1不仅优于完全微调的llm,而且具有优越的域内泛化能力。此外,这项工作代表了对多任务bioner微调llm的首次探索。结论:尽管需要的计算资源显着减少,但NERLlama3.1优于使用全参数更新进行微调的llm。此外,与传统的预训练语言模型相比,它表现出了显著优越的领域内泛化能力。它的低资源需求、高性能和强泛化增强了它在各种临床BioNER任务中的适用性和实用性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
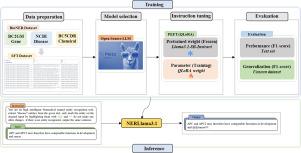
Resource-efficient instruction tuning of large language models for biomedical named entity recognition
Objective:
Large language models (LLMs) have exhibited remarkable efficacy in natural language processing (NLP) tasks, with fine-tuning for Biomedical Named Entity Recognition (BioNER) receiving significant research attention. However, the substantial computational demands associated with fine-tuning large-scale models constrain their development and deployment. Consequently, this study investigates parameter-efficient fine-tuning (PEFT) techniques to optimize LLMs for BioNER under limited computational resources. By leveraging these methods, competitive model performance is maintained while preserving in-domain generalization capability.
Methods:
In this study, we employed the PEFT method QLoRA to fine-tune the open-source Llama3.1 model, developing the NERLlama3.1 model specifically designed for the BioNER task. First, an LLM instruction tuning dataset was created using BioNER datasets such as NCBI-disease, BC5CDR-chem, and BC2GM-gene. Next, the Llama3.1-8B model was fine-tuned using the QLoRA method on a single 16GB memory GPU. Furthermore, during the inference phase, we introduced a prompt engineering technique called self-consistency NER prompting (SCNP). This approach leverages the diversity of outputs generated by LLMs to significantly enhance NER performance. Finally, we also developed a multi-task BioNER-capable model, NERLlama3.1-MT, to investigate the capability of fine-tuned LLMs in addressing multi-task BioNER scenarios.
Results:
The NERLlama3.1 model achieved F1-scores of 0.8977, 0.9402, and 0.8530 on the NCBI-disease, BC5CDR-chemical, and BG2GM-gene datasets, respectively. Furthermore, when evaluated on previously unseen datasets, it attained F1-scores of 0.6867 on BC5CDR-disease, 0.6800 on NLM-chemical, and 0.8378 on NLM-gene. These results demonstrate that NERLlama3.1 not only outperforms fully fine-tuned LLMs but also exhibits superior in-domain generalization capabilities when compared to the BERT-base model. Additionally, this work represents the first exploration of fine-tuning LLMs for multi-task BioNER.
Conclusion:
NERLlama3.1 outperformed LLMs fine-tuned with full parameter updates, despite requiring significantly fewer computational resources. Moreover, it exhibited substantially superior in-domain generalization capabilities compared to traditional pre-trained language models. Its low resource demands, high performance, and strong generalization enhance its applicability and utility across diverse clinical BioNER tasks.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Biomedical Informatics
医学-计算机:跨学科应用
CiteScore
8.90
自引率
6.70%
发文量
243
审稿时长
32 days
期刊介绍:
The Journal of Biomedical Informatics reflects a commitment to high-quality original research papers, reviews, and commentaries in the area of biomedical informatics methodology. Although we publish articles motivated by applications in the biomedical sciences (for example, clinical medicine, health care, population health, and translational bioinformatics), the journal emphasizes reports of new methodologies and techniques that have general applicability and that form the basis for the evolving science of biomedical informatics. Articles on medical devices; evaluations of implemented systems (including clinical trials of information technologies); or papers that provide insight into a biological process, a specific disease, or treatment options would generally be more suitable for publication in other venues. Papers on applications of signal processing and image analysis are often more suitable for biomedical engineering journals or other informatics journals, although we do publish papers that emphasize the information management and knowledge representation/modeling issues that arise in the storage and use of biological signals and images. System descriptions are welcome if they illustrate and substantiate the underlying methodology that is the principal focus of the report and an effort is made to address the generalizability and/or range of application of that methodology. Note also that, given the international nature of JBI, papers that deal with specific languages other than English, or with country-specific health systems or approaches, are acceptable for JBI only if they offer generalizable lessons that are relevant to the broad JBI readership, regardless of their country, language, culture, or health system.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


