TargetPair:临床试验衍生的癌症治疗靶标组合的单细胞组学资源
IF 4.5
2区 生物学
Q1 BIOCHEMISTRY & MOLECULAR BIOLOGY
引用次数: 0
摘要
肿瘤微环境(TME)的复杂性使癌症治疗具有挑战性。多靶点策略通常表现出优越的临床效益,并主导着正在进行的癌症临床试验。然而,现有的多目标策略在很大程度上依赖于实证方法。多靶点肿瘤治疗开发中靶对的合理设计是当务之急,但仍然是一个重大挑战,这可能受益于单细胞组学技术来解码TME。然而,目前尚未对临床相关的肿瘤TPs及其单细胞水平的表征进行深入的调查。在这里,我们建立了TargetPair数据库,通过人工标注已经评估过抗肿瘤疗效的药物组合和多靶点药物的tp,并计算它们在单细胞水平上的组学特征,来解决这一空白。TargetPair数据库提供(1)从3,470个临床试验和1,396份出版物中手动注释的60个已批准的,1,580个临床期和7,424个实验期tp,以及(2)从55篇文献(包括3,022,556个单细胞,13种细胞类型,16种癌症类型和588名患者)中手动整理的scRNA-seq数据集中获得tp的组学特征(共表达,分布,表达细胞组分,途径和单细胞基因网络)。它可以通过多种搜索和浏览模式在https://www.targetpair.aiddlab.com上自由访问。本文章由计算机程序翻译,如有差异,请以英文原文为准。
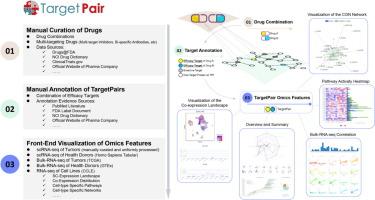
TargetPair: A Single Cell-omics Resource of Clinical Trials-derived Therapeutical Target Combinations for Cancer Therapy
The complexity of the tumor microenvironment (TME) makes cancer therapy challenging. Multi-targeting strategies often exhibit superior clinical benefits and dominate ongoing cancer clinical trials. However, existing multi-targeting strategies largely rely on empirical approaches. The rational design of target pairs (TPs) for multi-target cancer therapy development is a high priority but remains a significant challenge, which may benefit from single-cell omics technologies to decode the TME. However, there has been no thorough survey of clinically relevant cancer TPs and their characterization at the single-cell level. Here, we established the TargetPair database to address this gap by manually annotating TPs from drug combinations and multi-targeting drugs whose anti-tumor efficacy has been evaluated, and calculating their omics features at the single-cell level. The TargetPair database provides (1) 60 approved, 1,580 clinical-stage, and 7,424 experimental-stage TPs manually annotated from 3,470 clinical trials and 1,396 publications, and (2) the omics features of TPs (co-expression, distribution, expressed cell fractions, pathways, and single-cell gene networks) from manually curated scRNA-seq datasets derived from 55 pieces of literature (including 3,022,556 single cells, 13 cell types, 16 cancer types, and 588 patients). It can be freely accessed at https://www.targetpair.aiddlab.com via several search and browsing modes.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of Molecular Biology
生物-生化与分子生物学
CiteScore
11.30
自引率
1.80%
发文量
412
审稿时长
28 days
期刊介绍:
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology. The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and computational approaches to address challenging biological questions.
Research areas include but are not limited to: Biomolecular interactions, signaling networks, systems biology; Cell cycle, cell growth, cell differentiation; Cell death, autophagy; Cell signaling and regulation; Chemical biology; Computational biology, in combination with experimental studies; DNA replication, repair, and recombination; Development, regenerative biology, mechanistic and functional studies of stem cells; Epigenetics, chromatin structure and function; Gene expression; Membrane processes, cell surface proteins and cell-cell interactions; Methodological advances, both experimental and theoretical, including databases; Microbiology, virology, and interactions with the host or environment; Microbiota mechanistic and functional studies; Nuclear organization; Post-translational modifications, proteomics; Processing and function of biologically important macromolecules and complexes; Molecular basis of disease; RNA processing, structure and functions of non-coding RNAs, transcription; Sorting, spatiotemporal organization, trafficking; Structural biology; Synthetic biology; Translation, protein folding, chaperones, protein degradation and quality control.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


