scUCAF:用于单细胞多组学数据聚类的不确定性感知跨组学对齐和融合网络
IF 3.1
4区 生物学
Q2 BIOLOGY
引用次数: 0
摘要
单细胞多组学测序技术的发展为研究细胞异质性提供了新的思路。细胞聚类是多组学数据分析的关键步骤。然而,现有的方法往往忽略了组学中数据质量的变化,导致特征表示不可靠。为了解决这个问题,我们提出了一种用于多组学聚类的不确定性感知网络scUCAF。具体来说,为了减轻噪声对细胞特征提取的影响,我们引入了一个负二项分布的变分自编码器。在提取每个组学特征后,我们提出了一种高置信度的聚类引导对比学习方法,以确保跨组学特征的一致性。最后,一个不确定性感知融合和门控网络动态集成组学特征,以减轻低质量数据的偏差,并为聚类产生可靠的细胞表示。对8个真实单细胞多组学数据集的聚类结果表明,scUCAF聚类方法优于现有的多组学聚类方法。我们还进行了下游分析,以验证scUCAF在肝癌细胞类型注释和生物标志物鉴定中的有效性。本文章由计算机程序翻译,如有差异,请以英文原文为准。
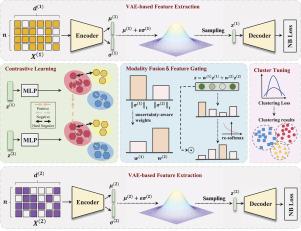
scUCAF: An uncertainty-aware cross-omics alignment and fusion network for single-cell multi-omics data clustering
The development of single-cell multi-omics sequencing technologies provides new insights into cell heterogeneity. Cell clustering is a crucial step in the analysis of multi-omics data. However, existing methods often overlook variations in data quality across omics, leading to unreliable feature representations. To address this issue, we propose scUCAF, an uncertainty-aware network for multi-omics clustering. Specifically, to mitigate the impact of noise on cell feature extraction, we introduce a variational autoencoder with a negative binomial distribution. After extracting each omics feature, we propose a high-confidence cluster-guided contrastive learning method to ensure cross-omics feature consistency. Finally, an uncertainty-aware fusion and gating network dynamically integrates the omics features to mitigate biases from low-quality data and produce reliable cell representations for clustering. Clustering results on eight real single-cell multi-omics datasets demonstrate that scUCAF outperforms existing multi-omics clustering methods. We also conduct downstream analyses to validate the effectiveness of scUCAF for cell type annotation and biomarker identification in liver cancer.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computational Biology and Chemistry
生物-计算机:跨学科应用
CiteScore
6.10
自引率
3.20%
发文量
142
审稿时长
24 days
期刊介绍:
Computational Biology and Chemistry publishes original research papers and review articles in all areas of computational life sciences. High quality research contributions with a major computational component in the areas of nucleic acid and protein sequence research, molecular evolution, molecular genetics (functional genomics and proteomics), theory and practice of either biology-specific or chemical-biology-specific modeling, and structural biology of nucleic acids and proteins are particularly welcome. Exceptionally high quality research work in bioinformatics, systems biology, ecology, computational pharmacology, metabolism, biomedical engineering, epidemiology, and statistical genetics will also be considered.
Given their inherent uncertainty, protein modeling and molecular docking studies should be thoroughly validated. In the absence of experimental results for validation, the use of molecular dynamics simulations along with detailed free energy calculations, for example, should be used as complementary techniques to support the major conclusions. Submissions of premature modeling exercises without additional biological insights will not be considered.
Review articles will generally be commissioned by the editors and should not be submitted to the journal without explicit invitation. However prospective authors are welcome to send a brief (one to three pages) synopsis, which will be evaluated by the editors.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


