探索LRRK2/PP1CA干扰肽与PP1CA的相互作用
IF 3
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
帕金森病(PD)是一种复杂的神经退行性疾病,目前只有姑息治疗。最近,亮氨酸到达重复激酶2 (LRRK2)和蛋白磷酸酶1 (PP1)之间的相互作用作为PD的优先靶点已经被发现,但LRRK2和PP1相互作用的方式目前尚不清楚,阻碍了调节剂的设计。我们之前已经发现了一个LRRK2片段能够干扰LRRK2和PP1催化亚基α (PP1CA)的相互作用,我们认为它应该在LRRK2/PP1CA界面上与PP1CA结合。在本研究中,我们首先鉴定了PP1CA中能够干扰LRRK2/PP1CA相互作用的片段,从而获得PP1CA可能面对LRRK2的区域信息。接下来,通过将计算机研究与体外竞争实验相结合,我们缩小了关于其与PP1CA结合模式的各种假设。总体而言,我们确定了LRRK2片段与PP1CA之间的一种首选结合模式,与初始片段突变体的体外结果一致。这些结果为进一步合理设计LRRK2/PP1CA相互作用的调节剂,进一步破译PD病因的分子机制铺平了道路。本文章由计算机程序翻译,如有差异,请以英文原文为准。
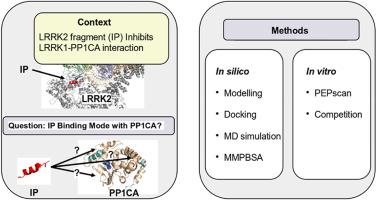
Exploring the interaction between a LRRK2/PP1CA interfering peptide and PP1CA
Parkinson's disease (PD) is a complex neurodegenerative disorder for which there is presently only palliative treatments. Recently, the interaction between the Leucine Reach Repeat Kinase 2 (LRRK2) and the Protein Phosphatase 1 (PP1) has come to light as a priority target for PD, but the way LRRK2 and PP1 interact is currently unknown, hampering the design of modulators. We have previously identified a fragment of LRRK2 able to interfere with the interaction of LRRK2 and PP1 catalytic subunit alpha (PP1CA), and we assume it should bind PP1CA at the LRRK2/PP1CA interface. In this study, we first identify the fragments of PP1CA that are able to interfere with the LRRK2/PP1CA interaction so as to get information about the regions of PP1CA likely to face LRRK2. Next, by combining in silico studies with in vitro competition experiments, we narrow down among the various hypotheses proposed regarding its mode of binding to PP1CA. Overall, we identify one preferred binding mode between the LRRK2 fragment and PP1CA consistent with in vitro results obtained for mutants of the initial fragment. These results pave the way for further rational design of modulators of the LRRK2/PP1CA interaction, to further decipher the molecular mechanisms underlying PD etiology.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


