GIPR同型二聚化的综合计算模型及相互作用机制
IF 3
4区 生物学
Q2 BIOCHEMICAL RESEARCH METHODS
引用次数: 0
摘要
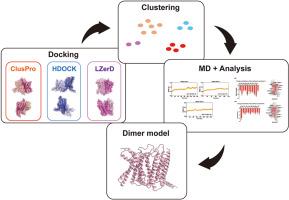
G蛋白偶联受体(gpcr)是一种跨膜受体,通过与G蛋白和其他效应器相互作用来调节细胞内信号传导,影响各种生理过程。葡萄糖依赖型胰岛素性多肽受体(GIPR)是GPCR家族的B1类成员,由GIP激活,通过增加葡萄糖依赖型胰岛素分泌、延缓胃排空、抑制食欲来调节餐后血糖。最近的研究强调跨膜结构域(TMD)是二聚化的主要界面,使GPCR形成具有不同生理作用的同型二聚体或异源二聚体。然而,这些二聚体的瞬态性质给结构分析带来了挑战,阻碍了实验探索和药物开发。计算方法现在为预测这种相互作用提供了强大的工具。本研究采用混合方法,结合多蛋白对接软件和动态结构优化,生成GIPR-TMD的潜在同二聚体模型。此外,下一步,验证模型将提供二聚体激活机制的见解,并支持新的治疗发现。本文章由计算机程序翻译,如有差异,请以英文原文为准。
Comprehensive computational modeling and interactions mechanisms of GIPR homodimerization
G protein-coupled receptors (GPCRs) are transmembrane receptors that regulate intracellular signaling by interacting with G proteins and other effectors, influencing various physiological processes. The Glucose-dependent Insulinotropic Polypeptide Receptor (GIPR), a class B1 GPCR family member activated by GIP, regulates postprandial glycaemia by augmenting glucose-dependent insulin secretion, delaying gastric emptying, and suppressing appetite. Recent studies highlight the transmembrane domain (TMD) as the primary interface for dimerization, allowing GPCR to form homodimers or heterodimers with distinct physiological roles. However, the transient nature of these dimers challenges structural analysis, hindering experimental exploration and drug development. Computational methods now offer powerful tools for predicting such interactions. This study employs a hybrid approach, combining multiple protein docking software and dynamic structural optimization, to generate potential homodimeric models of GIPR-TMD. In addition, Next, validated models will provide insights into dimer activation mechanisms and support novel therapeutic discoveries.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Journal of molecular graphics & modelling
生物-计算机:跨学科应用
CiteScore
5.50
自引率
6.90%
发文量
216
审稿时长
35 days
期刊介绍:
The Journal of Molecular Graphics and Modelling is devoted to the publication of papers on the uses of computers in theoretical investigations of molecular structure, function, interaction, and design. The scope of the journal includes all aspects of molecular modeling and computational chemistry, including, for instance, the study of molecular shape and properties, molecular simulations, protein and polymer engineering, drug design, materials design, structure-activity and structure-property relationships, database mining, and compound library design.
As a primary research journal, JMGM seeks to bring new knowledge to the attention of our readers. As such, submissions to the journal need to not only report results, but must draw conclusions and explore implications of the work presented. Authors are strongly encouraged to bear this in mind when preparing manuscripts. Routine applications of standard modelling approaches, providing only very limited new scientific insight, will not meet our criteria for publication. Reproducibility of reported calculations is an important issue. Wherever possible, we urge authors to enhance their papers with Supplementary Data, for example, in QSAR studies machine-readable versions of molecular datasets or in the development of new force-field parameters versions of the topology and force field parameter files. Routine applications of existing methods that do not lead to genuinely new insight will not be considered.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


