LYnet:利用卷积和循环神经网络计算识别肿瘤T细胞抗原
IF 3.1
4区 生物学
Q2 BIOLOGY
引用次数: 0
摘要
免疫疗法代表了肿瘤学的范式转变,在疗效和特异性方面优于传统疗法。其成功的关键是识别t细胞抗原,这是触发有效的抗肿瘤免疫反应所必需的。然而,目前的抗原预测方法缺乏最佳疫苗开发所需的精度。本研究旨在通过引入一种新的深度学习模型来准确预测肿瘤t细胞抗原,从而解决这一空白。它力求改进鉴定过程,从而促进研制更有效的治疗性癌症疫苗。方法将一维卷积神经网络与双向长短期记忆层相结合,构建LYnet混合结构,同时捕获局部基序模式和长期序列依赖关系。19个互补的序列衍生描述符(包括aindex、AAK-mer、CKSAAP/CKSAAGP和理化成分向量)被连接起来形成输入特征空间。采用SMOTE-Tomek重采样策略减轻了训练集中的类不平衡。通过分层10倍交叉验证来量化模型的稳健性,并在两个独立的基准集合(TAP 1.0和iTTCA-RF)上验证泛化。结果在LYnet基准上进行10次验证,该模型的AUC为0.992,灵敏度为0.863,特异性为0.925,MCC为0.776。独立评估证实了该方法的优势:LYnet在TAP 1.0集上的AUC为0.949,在iTTCA-RF集上的AUC为0.836,在AUC上超过最强竞争方法2.4-6.9个百分点,在MCC上超过10.6个百分点。这些结果表明LYnet在肿瘤t细胞抗原鉴定中达到了最先进的准确性和平衡预测。结论该深度学习模型的引入在肿瘤t细胞抗原预测方面取得了重大进展。其优越的准确性和稳健性为促进癌症免疫治疗的发展和疗效提供了巨大的潜力。这项工作不仅强调了精确抗原识别在免疫治疗中的重要性,而且为帮助设计下一代癌症疫苗提供了强大的计算工具。本文章由计算机程序翻译,如有差异,请以英文原文为准。
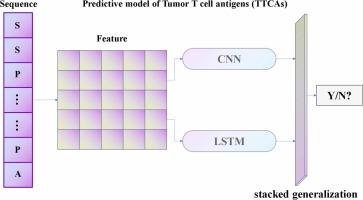
LYnet: Computational identification of tumor T cell antigens using convolutional and recurrent neural networks
Background
Immunotherapy represents a paradigm shift in oncology, offering advantages in efficacy and specificity over traditional therapies. Key to its success is the identification of T-cell antigens, which are essential for triggering an effective antitumor immune response. Current methodologies for antigen prediction, however, lack the precision required for optimal vaccine development.
Purpose
This study aims to address this gap by introducing a novel deep learning model for the accurate prediction of tumor T-cell antigens. It seeks to improve the identification process, thereby facilitating the creation of more effective therapeutic cancer vaccines.
Methods
A hybrid architecture, designated LYnet, was constructed by integrating one-dimensional Convolutional Neural Networks with bidirectional Long Short-Term Memory layers, thereby capturing both local motif patterns and long-range sequence dependencies. Nineteen complementary sequence-derived descriptors—including AAindex, AAK-mer, CKSAAP/CKSAAGP, and physicochemical composition vectors—were concatenated to form the input feature space. Class imbalance in the training set was mitigated with the SMOTE-Tomek resampling strategy. Model robustness was quantified by stratified 10-fold cross-validation, and generalisation was verified on two independent benchmark collections (TAP 1.0 and iTTCA-RF).
Results
Across 10-fold validation on the LYnet benchmark, the proposed model achieved an AUC of 0.992, together with a sensitivity of 0.863, specificity of 0.925 and MCC of 0.776. Independent evaluation confirmed the advantage: LYnet yielded AUCs of 0.949 on the TAP 1.0 set and 0.836 on the iTTCA-RF set, surpassing the strongest competing method by 2.4–6.9 percentage points in AUC and up to 10.6 percentage points in MCC. These results demonstrate that LYnet attains state-of-the-art accuracy and balanced prediction for tumour T-cell antigen identification.
Conclusions
The introduction of this deep learning model represents a significant advancement in the prediction of tumor T-cell antigens. Its superior accuracy and robustness offer substantial potential to enhance the development and efficacy of cancer immunotherapies. This work not only underscores the importance of precise antigen identification in immunotherapy but also provides a powerful computational tool to aid in the design of next-generation cancer vaccines.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Computational Biology and Chemistry
生物-计算机:跨学科应用
CiteScore
6.10
自引率
3.20%
发文量
142
审稿时长
24 days
期刊介绍:
Computational Biology and Chemistry publishes original research papers and review articles in all areas of computational life sciences. High quality research contributions with a major computational component in the areas of nucleic acid and protein sequence research, molecular evolution, molecular genetics (functional genomics and proteomics), theory and practice of either biology-specific or chemical-biology-specific modeling, and structural biology of nucleic acids and proteins are particularly welcome. Exceptionally high quality research work in bioinformatics, systems biology, ecology, computational pharmacology, metabolism, biomedical engineering, epidemiology, and statistical genetics will also be considered.
Given their inherent uncertainty, protein modeling and molecular docking studies should be thoroughly validated. In the absence of experimental results for validation, the use of molecular dynamics simulations along with detailed free energy calculations, for example, should be used as complementary techniques to support the major conclusions. Submissions of premature modeling exercises without additional biological insights will not be considered.
Review articles will generally be commissioned by the editors and should not be submitted to the journal without explicit invitation. However prospective authors are welcome to send a brief (one to three pages) synopsis, which will be evaluated by the editors.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


