完成基因表达预测基准的空间转录组学数据
IF 11.8
1区 医学
Q1 COMPUTER SCIENCE, ARTIFICIAL INTELLIGENCE
引用次数: 0
摘要
空间转录组学是一项开创性的技术,将组织学图像与空间分辨率的基因表达谱相结合。在各种可用的空间转录组学技术中,Visium已成为最广泛采用的技术。然而,其可及性受到成本高、需要专业知识和临床整合缓慢等因素的限制。此外,基因捕获效率低下导致严重的辍学,破坏了获得的数据。为了应对这些挑战,深度学习社区已经探索了直接从组织学图像中进行基因表达预测的任务。然而,数据集、预处理和训练协议的不一致性阻碍了模型之间的公平比较。为了弥补这一差距,我们引入了spare,这是一个由26个公共数据集组成的系统策划数据库,为模型评估提供了标准化资源。我们进一步提出了SpaCKLE,这是一种最先进的基于转换器的基因表达完成模型,与现有方法相比,其均方误差降低了82.5%以上。最后,我们建立了spare基准,在原始数据和SpaCKLE完成的数据上评估了8个最先进的预测模型,表明SpaCKLE在所有基因表达预测模型上都大大提高了结果。总之,我们的贡献构成了迄今为止从组织学图像中预测基因表达的最全面的基准,并为未来空间转录组学研究奠定了基础。本文章由计算机程序翻译,如有差异,请以英文原文为准。
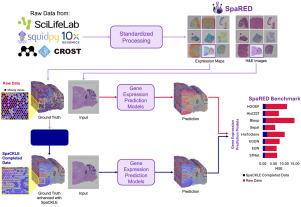

Completing spatial transcriptomics data for gene expression prediction benchmarking
Spatial Transcriptomics is a groundbreaking technology that integrates histology images with spatially resolved gene expression profiles. Among the various Spatial Transcriptomics techniques available, Visium has emerged as the most widely adopted. However, its accessibility is limited by high costs, the need for specialized expertise, and slow clinical integration. Additionally, gene capture inefficiencies lead to significant dropout, corrupting acquired data. To address these challenges, the deep learning community has explored the gene expression prediction task directly from histology images. Yet, inconsistencies in datasets, preprocessing, and training protocols hinder fair comparisons between models. To bridge this gap, we introduce SpaRED, a systematically curated database comprising 26 public datasets, providing a standardized resource for model evaluation. We further propose SpaCKLE, a state-of-the-art transformer-based gene expression completion model that reduces mean squared error by over 82.5% compared to existing approaches. Finally, we establish the SpaRED benchmark, evaluating eight state-of-the-art prediction models on both raw and SpaCKLE-completed data, demonstrating SpaCKLE substantially improves the results across all the gene expression prediction models. Altogether, our contributions constitute the most comprehensive benchmark of gene expression prediction from histology images to date and a stepping stone for future research on Spatial Transcriptomics.
求助全文
通过发布文献求助,成功后即可免费获取论文全文。
去求助
来源期刊

Medical image analysis
工程技术-工程:生物医学
CiteScore
22.10
自引率
6.40%
发文量
309
审稿时长
6.6 months
期刊介绍:
Medical Image Analysis serves as a platform for sharing new research findings in the realm of medical and biological image analysis, with a focus on applications of computer vision, virtual reality, and robotics to biomedical imaging challenges. The journal prioritizes the publication of high-quality, original papers contributing to the fundamental science of processing, analyzing, and utilizing medical and biological images. It welcomes approaches utilizing biomedical image datasets across all spatial scales, from molecular/cellular imaging to tissue/organ imaging.
 求助内容:
求助内容: 应助结果提醒方式:
应助结果提醒方式:


